Abstract
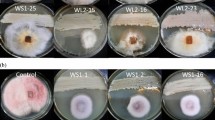
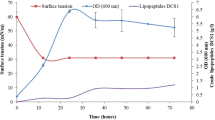
In this study, nine strains of Pseudomonas aurantiaca and P. chlororaphis, and two isolates of Pseudomonas sp.: At1RP4 and RS-1, were characterized for the in-vitro production of secondary metabolites in LB, DMB, and King’s B media, and of the genes responsible for the production of antagonistic metabolites. Based on 16S rRNA gene sequence, isolates At1RP4 and RS-1 were identified as strains of P. aeruginosa and P. fluorescens. Five phenazine derivatives comprising phenazine, phenazine-1-carboxylic acid (PCA), 2-hydroxyphenazine-1-carboxylic acid (2-OH-Phz-1-COOH), phenazine-1,6-dicarboxylic acid (Phz-1,6-di-COOH), and 2-hydroxyphenazine (2-OH-Phz) were produced by all strains in all three culture media including DMB, King’s B and LB. However, 2,8-dihydroxyphenazine, 6-methylphenazine-1-carboxylic acid, pyrrolnitrin, and the ortho-dialkyl-aromatic acids, were produced by the P. aurantiaca and P. chlororaphis strains. In addition, all strains produced 2-acetamidophenol, pyochelin, and diketopiperazine derivatives in variable amounts in all three culture media used. Highest levels of quorum-sensing signal molecules including PQS, 2-Octyl-3-hydroxy-4(1H)-quinolone, and hexahydro-quinoxaline-1,4-dioxide were recorded for P. aeruginosa At1RP4. Moreover, all strains produced volatile hydrogen cyanide (0.95–6.68 µg/L) and the phytohormone indole-3-acetic acid (0.42–13.9 µM). Production of extracellular lipase and protease was recorded in all pseudomonads, whereas, cellulase production and phosphate solubilization were variable. Genes for hydrogen cyanide and phenazine-1-carboxylic acid were detected in all eleven strains while the gene for pyrrolnitrin biosynthesis was amplified in P. aurantiaca and P. chlororaphis strains. Comparative metabolomic analysis provided detailed insights about the strain-specific metabolites in pseudomonads, and their pseudo-relative quantification in different bacterial growth media to be used as single-strain biofertilizer and biocontrol inoculums.





Similar content being viewed by others
References
Allen F, Pon A, Wilson M, Greiner R, Wishart D (2014) CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acid Res 42:W94-99
Alnahdi HS (2012) Isolation and screening of extracellular proteases produced by new isolated Bacillus sp. JAPS 2:71–74
Bauer JS, Hauck N, Christof L, Mehnaz S, Gust B, Gross H (2016) The systematic investigation of the quorum sensing system of the biocontrol strain Pseudomonas chlororaphis subsp. aurantiaca PB-St2 unveils aurI to be a biosynthetic origin for 3-oxo-homoserine lactones. PLoS ONE 11:0167002
Bertani G (2004) Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186:595–600
Cezairliyan B, Vinayavekhin N, Grenfell-Lee D, Yuen GJ, Saghatelian A, Ausubel FM (2013) Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1003101
Chin-A-Woeng TFC, Bloemberg GV, Van-der-Bij AJ, Van-der-Drift KMGM, Schripsema J, Kroon B, Scheffer RJ, Keel C, Bakker PAHM, Tichy H, de Bruijn FJ, Thomas-Oates JE, Lugtenberg BJJ (1998) Biocontrol by phenazine-1-carboxamide producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. MPMI 11:1069–1077
Compant S, Reiter B, Sessitsch A, Nowak J, Clément C, Barka EA (2005) Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. Strain PsJN Appl Environ Microbiol 71:1685–1693
Davis BD (1949) The isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc Natl Acad Sci 35:1–10
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465-469
De-Weger LA, Van-der Bij AJ, Dekkers LC, Simons M, Wijffelman CA, Lugtenberg BJ (1995) Colonization of the rhizosphere of crop plants by plant-beneficial pseudomonads. FEMS Microbiol Ecol 17:221–227
Diggle SP, Cornelis P, Williams P, Camara M (2006) 4-Quinolone signaling in Pseudomonas aeruginosa: old molecules, new perspectives. Int J Med Microbiol 296:83–91
Goswami D, Thakker JN, Dhandhukia PC (2015) Simultaneous detection and quantification of indole-3-acetic acid (IAA) and indole-3-butyric acid (IBA) produced by rhizobacteria from L-tryptophan (Trp) using HPTLC. J Microbiol Methods 110:7–14
Hayat R, Ali S, Amara U, Khalid R, Ahmed I (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 60:579–598
Huang L, Chen MM, Wang W, Hu HB, Peng HS, Xu YQ, Zhang XH (2011) Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Applied Microbiol Biotechnol 89:169–177
Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A (2008) A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Current Microbiol 57:503–507
Kim JS, Pauline MM, David EC (2013) Application of PCR primer sets for detection of Pseudomonas sp. functional genes in the plant rhizosphere. J Agric Chem Environ 2:8–15
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med 44:301–307
Kirner S, Hammer PE, Hill DS, Altmann A, Fischer I, Weislo LJ, Lanahan M, van Pee KH, Ligon JM (1998) Functions encoded by pyrrolnitrin biosynthetic genes from Pseudomonas fluorescens. J Bacteriol 180:1939–1943
Kraus J, Loper JR (1995) Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 61:849–854
Kumar RS, Ayyadurai N, Pandiaraja P, Reddy AV, Venkateswarlu Y, Prakash O, Sakthivel N (2005) Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity and biofertilizing traits. J Appl Microbiol 98:145–154
Lambert JL, Ramasamy J, Paukstelis JV (1975) Stable reagents for the colorimetric determination of cyanide by modified Konig reactions. Anal Chem 47:916–918
Ligon JM, Hill DS, Hammer PE, Torkewitz NR, Hofmann D, Kempf HJ, van-Pee KH (2000) Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag Sci 56:688–695
Loeschcke A, Thies S (2015) Pseudomonas putida-a versatile host for the production of natural products. Appl Microbiol Biotechnol 99:6197–6214
Matthijs S, Budzikiewicz H, Schafer M, Wathelet B, Cornelis B (2008) Ornicorrugatin, a new siderophore from Pseudomonas fluorescens AF76. Z Naturforsch 63:8–12
Mehnaz S, Saleem RSZ, Yameen B, Pianet I, Schnakenburg G, Pietraszkiewicz H, Valeriote F, Josten M, Sahl HG, Franzblau SG, Gross H (2013) Lahorenoic acids A-C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. J Nat Prod 76:135–141
Mehnaz S, Bauer JS, Gross H (2014) Complete genome sequence of the sugar cane endophyte Pseudomonas aurantiaca PB-St2, a disease-suppressive bacterium with antifungal activity toward the plant pathogen Colletotrichum falcatum. Genome Announc 2:e1108–e1113. https://doi.org/10.1128/genomeA.01108-13
Millar R, Higgins VJ (1970) Association of cyanide with infection of bird’s foot trefoil by Stemphylium loti. Phytopathology 60:104–110
Minagawa S, Inami H, Kato T, Sawada S, Yasuki T, Miyairi S, Horikawa M, Okuda J, Gotoh N (2012) RND type efflux pump system MexAB-OprM of Pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol. https://doi.org/10.1186/1471-2180-12-70
Morrison CK, Arseneault T, Novinscak A, Filio M (2017) Phenazine-1-carboxylic acid production by Pseudomonas fluorescens LBUM636 alters Phytophthora infestans growth and late blight development. Phytopathology 107:273–279
Naik PR, Sakthivel N (2006) Functional characterization of a novel hydrocarbonoclastic Pseudomonas sp. strain PUP6 with plant-growth-promoting traits and antifungal potential. Res Microbiol 157:538–546
Normand P (1995) Utilisation des séquences 16S pour le positionnement phylétique d’un organisme inconnu. Oceanis 21:31–56
Oteino N, Lally RD, Kiwanuka S, Lloyd A, Ryan D, Germaine KJ, Dowling DN (2015) Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front Microbiol. https://doi.org/10.3389/fmicb.2015.00745
Park JY, Oh SA, Anderson AJ, Neiswender J, Kim JC, Kim YC (2011) Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose. Lett Appl Microbiol 52:532–537
Pathma J, Ayyadurai N, Sakthivel N (2010) Assessment of genetic and functional relationship of antagonistic fluorescent pseudomonads of rice rhizosphere by repetitive sequence, protein coding sequence and functional gene analyses. J Microbiol 48:715–727
Pikovskaya R (1948) Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17:e370
Saharan B, Nehra V (2011) Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res 21. http://astonjournals.com/lsmr
Schwyn B, Neilands J (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Selin C, Habibian R, Poritsanos N, Athukorala SN, Fernando D, de Kievit TR (2010) Phenazines are not essential for Pseudomonas chlororaphis PA23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol Ecol 71:73–83
Shahid I, Rizwan M, Baig DN, Saleem RS, Malik KA, Mehnaz S (2017) Secondary metabolites production and plant growth promotion by Pseudomonas chlororaphis subsp. aurantiaca strains isolated from cotton, cactus, and para grass. J Microbiol Biotechnol 27:480–491
Sierra G (1957) A simple method for the detection of lipolytic activity of micro-648 organisms and some observations on the influence of the contact between cells and fatty 649 acid substrates. Antonie Leeuwenhoek 23:15–22
Smaoui S, Mellouli L, Lebrihi A, Coppel Y, Fguira LFB, Mathieu F (2011) Purification and structure elucidation of three naturally bioactive molecules from the new terrestrial Streptomyces sp. TN17 strain. Nat Prod Res 25:806–814
Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27:747–751
Tashiro Y, Yawata Y, Toyofuku M, Uchiyama H, Nomura N (2013) Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ 28:13–24
Thongsri Y, Aromdee C, Yenjai C, Kanokmedhakul S, Chaiprasert A, Prariyachatigul C (2014) Detection of diketopiperazine and pyrrolnitrin, compounds with anti-Pythium insidiosum activity, in a Pseudomonas stutzeri environmental strain. Biomed Pap 158:378–383
Vázquez RD, González O, Guzmán-Rodríguez J, Perez ALD, Zaarzosa AO, Bucio JL, Carmen VM, Garcia JC (2015) Cytotoxicity of cyclodipeptides from Pseudomonas aeruginosa PAO1 leads to apoptosis in human cancer cell lines. BioMed Res Intern. https://doi.org/10.1155/2015/197608
Wang M et al (2016) Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol 34:828–837
Welbaum GE, Sturz AV, Dong Z, Nowak J (2004) Managing soil microorganisms to improve productivity of agroecosystems. Crit Rev Plant Sci 23:175–193
Yasmin S, Hafeez FY, Mirza MS, Rasul M, Arshad HMI, Zubair M, Iqbal M (2017) Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front Microbiol 8:1895. https://doi.org/10.3389/fmicb.2017.01895
Yu JM, Wang D, Pierson LS III, Pierson EA (2018) Effect of producing different phenazines on bacterial fitness and biological control in Pseudomonas chlororaphis 30–84. Plant Pathol J 34:44
Zhang Y, Fernando WG, Kievit TRD, Berry C, Daayf F, Paulitz T (2006) Detection of antibiotic-related genes from bacterial biocontrol agents with polymerase chain reaction. Can J Microbiol 52:476–481
Acknowledgements
This work received financial support from Higher Education Commission (HEC) of Pakistan. The funding was awarded to Izzah Shahid for PhD research work. The metabolomics research at the UVic-Genome BC Proteomics Centre was supported by funding to “The Metabolomics Innovation Centre (TMIC)” through the Genome Innovations Network (GIN) from Genome Canada, Genome Alberta, and Genome British Columbia for operations (205MET and 7203) and for technology development (215MET and MC3T) in metabolomics. The authors would like to thank Dr. Carol E. Parker for assistance with English language editing.
Author information
Authors and Affiliations
Contributions
IS performed experiments and prepared manuscript initially. CHB and JH provided the conceptual framework for the experiments. DNB helped in genetic analysis. DH assisted in LC–MS and data analysis. SM supervised the research as well as feedback and guidance during manuscript development. KAM managed resources and critically read the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shahid, I., Han, J., Hardie, D. et al. Profiling of antimicrobial metabolites of plant growth promoting Pseudomonas spp. isolated from different plant hosts. 3 Biotech 11, 48 (2021). https://doi.org/10.1007/s13205-020-02585-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02585-8