Abstract
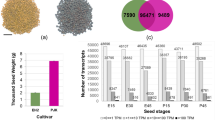
Grain size is an important trait for crops. The endogenous hormones brassinosteroids (BRs) play key roles in grain size and mass. In this study, we identified an ethyl methylsulfonate (EMS) mutant wheat line, SM482gs, with increased grain size, 1000-grain weight, and protein content, but decreased starch content, compared with the levels in the wild type (WT). Comparative transcriptomic analysis of SM482gs and WT at four developmental stages [9, 15, 20, and 25 days post-anthesis (DPA)] revealed a total of 264, 267, 771, and 1038 differentially expressed genes (DEGs) at these stages. Kyoto Encyclopedia of Genes and Genomes (KEGG) database analysis showed that some DEGs from the comparison at 15 DPA were involved in the pathway of “brassinosteroid biosynthesis,” and eight genes involved in BR biosynthesis and signal transduction were significantly upregulated in SM482gs during at least one stage. This indicated that the enhanced BR signaling in SM482gs might have contributed to its increased grain size via network interactions. The expression of seed storage protein (SSP)-encoding genes in SM482gs was upregulated, mostly at 15 and 20 DPA, while most of the starch synthetase genes showed lower expression in SM482gs at all stages, compared with that in WT. The expression patterns of starch synthase genes and seed storage protein-encoding genes paralleled the decreased level of starch and increased storage protein content of SM482gs, which might be related to the increased seed weight and wrinkled phenotype.







Similar content being viewed by others
Data availability
The raw sequencing data were deposited in the NCBI SRA database with accession numbers SRP115001/SRR5908629 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP115001).
Abbreviations
- BR:
-
Brassinosteroid
- EMS:
-
Ethyl methanesulfonate
- WT:
-
Wild type
- DPA:
-
Days post-anthesis
- DEG:
-
Differentially expressed gene
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- HMW-GS:
-
High-molecular-weight glutenin subunit
- LMW-GS:
-
Low-molecular-weight glutenin subunit
- RT-qPCR:
-
Real-time quantitative reverse transcription polymerase chain reaction
- RNA-Seq:
-
Ribonucleic acid sequencing
- TPM:
-
Transcripts per million
- AGPase:
-
Adenosine diphosphate glucose pyrophosphorylase
- SS:
-
Starch synthase
- SBE:
-
Starch branching enzyme
- DBE:
-
Starch debranching enzyme
- GBSS:
-
Granule-bound starch synthase
References
Abdoli M, Saeidi M, Jalali-Honarmand S, Mansourifar S, Ghobadi M-E, Cheghamirza K (2013) Effect of source and sink limitation on yield and some agronomic characteristics in modern bread wheat cultivars under post anthesis water deficiency. Acta agriculturae Slovenica 101 (2). doi:https://doi.org/10.2478/acas-2013-0013
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106. https://doi.org/10.1186/gb-2010-11-10-r106
Bajguz A (2007) Metabolism of brassinosteroids in plants. Plant Physiol Biochem 45(2):95–107. https://doi.org/10.1016/j.plaphy.2007.01.002
Bittner T, Campagne S, Neuhaus G et al (2013) Identification and characterization of two wheat Glycogen Synthase Kinase 3/SHAGGY-like kinase. BMC Plant Biol 13
Blanco A, Lotti C, Simeone R et al (2001) Detection of quantitative trait loci for grain yield and yield components across environments in durum wheat. Cereal Res Commun 29:237–244. https://doi.org/10.1534/genetics.105.044586
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172(2):1165–1177
Brown TA, Jones MK, Powell W, Allaby RG (2009) The complex origins of domesticated crops in the Fertile Crescent. Trends Ecol Evol 24(2):103–109. https://doi.org/10.1016/j.tree.2008.09.008
Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, Zhao M, Chu C (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nature plants 2:15195. https://doi.org/10.1038/nplants.2015.195
Choe S, Dilkes BP, Fujioka S et al (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that meditates multiple 22a-Hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10:231–243
Clavijo BJ, Venturini L, Schudoma C et al (2017) An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res 27(5):885–896. https://doi.org/10.1101/gr.217117.116
Clouse SD (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23(4):1219–1230. https://doi.org/10.1105/tpc.111.084475
Duan K, Li L, Hu P, Xu SP, Xu ZH, Xue HW (2006) A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J 47(4):519–531. https://doi.org/10.1111/j.1365-313X.2006.02804.x
Fujioka S, Li J, Choi YH et al (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Am Soc Plant Physiol 9:1951–1962
Gegas VC, Nazari A, Griffiths S, Simmonds J, Fish L, Orford S, Sayers L, Doonan JH, Snape JW (2010) A genetic framework for grain size and shape variation in wheat. Plant Cell 22(4):1046–1056. https://doi.org/10.1105/tpc.110.074153
Goetz M, Godt DE, Roitsch T (2000) Tissue-specific induction of the mRNA for an extracellular invertase isoenzyme of tomato by brassinosteroids suggests a role for steroid hormones in assimilate partitioning. Plant J 22(6):515–522. https://doi.org/10.1046/j.1365-313x.2000.00766.x
Guan Y, Li G, Chu Z, Ru Z, Jiang X, Wen Z, Zhang G, Wang Y, Zhang Y, Wei W (2019) Transcriptome analysis reveals important candidate genes involved in grain-size formation at the stage of grain enlargement in common wheat cultivar “Bainong 4199.” PLoS ONE 14(3):e0214149. https://doi.org/10.1371/journal.pone.0214149
Gudesblat GE, Russinova E (2011) Plants grow on brassinosteroids. Curr Opin Plant Biol 14(5):530–537. https://doi.org/10.1016/j.pbi.2011.05.004
Hannah LC, James M (2008) The complexities of starch biosynthesis in cereal endosperms. Curr Opin Biotechnol 19(2):160–165. https://doi.org/10.1016/j.copbio.2008.02.013
Hauvermale AL, Ariizumi T, Steber CM (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160(1):83–92. https://doi.org/10.1104/pp.112.200956
He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99(15):10185–10190. https://doi.org/10.1073/pnas.152342599
Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) The Rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17(8):2243–2254. https://doi.org/10.1105/tpc.105.030973
Hong Y, Chen L, Du LP, Su Z, Wang J, Ye X, Qi L, Zhang Z (2014) Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct Integr Genomics 14(2):341–349. https://doi.org/10.1007/s10142-014-0380-5
Hoshino T, Ito S, Hatta K et al (1996) Development of waxy common wheat by haploid breeding. Breed Sci 46:185–188
Hush D, Scovel HC (2005) Concentration of the hypergeometric distribution. Stat Probabil Lett 2(75):127–132
Jenner C, Ugalde T, Aspinall D (1991) The physiology of starch and protein deposition in the endosperm of wheat. AustJPlant Physiol 18:211–226
Jiang WB, Huang HY, Hu YW, Zhu SW, Wang ZY, Lin WH (2013) Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol 162(4):1965–1977. https://doi.org/10.1104/pp.113.217703
Khatkar BS, Barak S, Mudgil D (2013) Effects of gliadin addition on the rheological, microscopic and thermal characteristics of wheat gluten. Int J Biol Macromol 53:38–41. https://doi.org/10.1016/j.ijbiomac.2012.11.002
Khew CY, Teo CJ, Chan WS, Wong HL, Namasivayam P, Ho CL (2015) Brassinosteroid insensitive 1-associated kinase 1 (OsI-BAK1) is associated with grain filling and leaf development in rice. J Plant Physiol 182:23–32. https://doi.org/10.1016/j.jplph.2015.05.003
Kim TW, Wang ZY (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol 61:681–704. https://doi.org/10.1146/annurev.arplant.043008.092057
Koh S, Lee SC, Kim MK, Koh JH, Lee S, An G, Choe S, Kim SR (2007) T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol Biol 65(4):453–466. https://doi.org/10.1007/s11103-007-9213-4
Kovalchuk N, Smith J, Pallotta M et al (2009) Characterization of a wheat endosperm transfer cell specific protein TaPR60. Plant Mol Biol 71:81–98. https://doi.org/10.1007/s11103-009-9510-1)
Lagrain B, Thewissen BG, Brijs K, Delcour JA (2008) Mechanism of gliadin–glutenin cross-linking during hydrothermal treatment. Food Chem 107(2):753–760. https://doi.org/10.1016/j.foodchem.2007.08.082
Langmead, B., Trapnell, C., Pop, M. et al (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10:R25 (3). doi:https://doi.org/10.1093/bioinformatics/btp120
Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295(5558):1299–1301. https://doi.org/10.1126/science.1065769
Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7(8):791–806. https://doi.org/10.1111/j.1467-7652.2009.00444.x
Liu S, Hua L, Dong S, Chen H, Zhu X, Jiang J, Zhang F, Li Y, Fang X, Chen F (2015) OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J 84(4):672–681. https://doi.org/10.1111/tpj.13025
Liu J, Chen J, Zheng X, Wu F, Lin Q, Heng Y, Tian P, Cheng Z, Yu X, Zhou K, Zhang X, Guo X, Wang J, Wang H, Wan J (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat Plants 3:17043. https://doi.org/10.1038/nplants.2017.43
Lu J, Chang C, Zhang HP, Wang SX, Sun G, Xiao SH, Ma CX (2015) Identification of a novel allele of TaCKX6a02 associated with grain size, filling rate and weight of common wheat. PLoS ONE 10(12):e0144765. https://doi.org/10.1371/journal.pone.0144765
Ma M, Wang Q, Li Z, Cheng H, Li Z, Liu X, Song W, Appels R, Zhao H (2015) Expression of TaCYP78A3, a gene encoding cytochrome P450 CYP78A3 protein in wheat (Triticum aestivum L.), affects seed size. The Plant journal: for cell and molecular biology 83(2):312–325. doi:https://doi.org/10.1111/tpj.12896
Ma L, Li T, Hao C, Wang Y, Chen X, Zhang X (2016) TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol J 14(5):1269–1280. https://doi.org/10.1111/pbi.12492
Mangalika WHA, Miura H, Yamauchi H et al (2003) Properties of starches from near-isogenic wheat lines with different Wx protein deficiencies. Cereal Chem 6(80):662–666
Melnyk JP, Dreisoerner J, Marcone MF, Seetharaman K (2012) Using the gluten peak tester as a tool to measure physical properties of gluten. J Cereal Sci 56(3):561–567. https://doi.org/10.1016/j.jcs.2012.07.015
Meyer FD, Smidansky ED, Beecher B, Greene TW, Giroux MJ (2004) The maize Sh2r6hs ADP-glucose pyrophosphorylase (AGP) large subunit confers enhanced AGP properties in transgenic wheat (Triticum aestivum). Plant Sci 167(4):899–911. https://doi.org/10.1016/j.plantsci.2004.05.031
Mishra A, Singh A, Sharma M, Kumar P, Roy J (2016) Development of EMS-induced mutation population for amylose and resistant starch variation in bread wheat (Triticum aestivum) and identification of candidate genes responsible for amylose variation. BMC Plant Biol 16(1):217. https://doi.org/10.1186/s12870-016-0896-z
Morinaka Y (2006) Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol 141(3):924–931. https://doi.org/10.1104/pp.106.077081
Mungall CJ, Bada M, Berardini TZ, Deegan J, Ireland A, Harris MA, Hill DP, Lomax J (2011) Cross-product extensions of the Gene Ontology. J Biomed Inform 44(1):80–86. https://doi.org/10.1016/j.jbi.2010.02.002
Nakamura T, Yamamori M, Hirano H, Hidaka S (1993) Identification of three Wx proteins in wheat (Triticum aestivum L.). Biochem Genet 31(1–2):75–86. https://doi.org/10.1007/BF02399821
Navarro C, Moore J, Ott A, Baumert E, Mohan A, Gill KS, Sandhu D (2015) Evolutionary, comparative and functional analyses of the brassinosteroid receptor gene, BRI1, in wheat and its relation to other plant genomes. PLoS ONE 10(5):e0127544. https://doi.org/10.1371/journal.pone.0127544
Nomura T, Ueno M, Yamada Y, Takatsuto S, Takeuchi Y, Yokota T (2007) Roles of brassinosteroids and related mRNAs in pea seed growth and germination. Plant Physiol 143(4):1680–1688. https://doi.org/10.1104/pp.106.093096
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27(1):29–34
Patil RM, Tamhankar SA, Oak MD et al (2013) Mapping of QTL for agronomic traits and kernel characters in durum wheat (Triticum durum Desf.). Euphytica 190:117–129. https://doi.org/10.1007/s10681-012-0785-y)
Pfeifer M, Kugler KG, Sandve SR et al (2014) Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345(6194):1250091–1250095. https://doi.org/10.1126/science.1251788
Rao HKS, Sears ER (1964) Chemical mutagenesis in Triticum aestivum. Mutation Res Fundamental Mol Mech Mutagenesis 1(4):387–399
Regina A, Berbezy P, Kosar-Hashemi B, Li S, Cmiel M, Larroque O, Bird AR, Swain SM, Cavanagh C, Jobling SA, Li Z, Morell M (2015) A genetic strategy generating wheat with very high amylose content. Plant Biotechnol J 13(9):1276–1286. https://doi.org/10.1111/pbi.12345
Robert HS, Grunewald W, Sauer M, Cannoot B, Soriano M, Swarup R, Weijers D, Bennett M, Boutilier K, Friml J (2015) Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142(4):702–711. https://doi.org/10.1242/dev.115832
Schulman AH, Tester RF, Ahokas H, Morrison WR (1994) The effect of the shrunken endosperm mutation shx on starch granule development in barley seeds. J Cereal Sci 19(1):49–55. https://doi.org/10.1006/jcrs.1994.1007
Shahidullah SM, Hanafi MM, Ashrafuzzaman M et al (2010) Analysis of grain density and yield characters in aromatic rice genotypes. Agrociencia 44(7):325–227
Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40(8):1023–1028. https://doi.org/10.1038/ng.169
Singh A, Breja P, Khurana JP, Khurana P (2016) Wheat brassinosteroid-insensitive1 (TaBRI1) interacts with members of TaSERK gene family and cause early flowering and seed yield enhancement in Arabidopsis. PLoS ONE 11(6):e0153273. https://doi.org/10.1371/journal.pone.0153273
Smidansky ED, Clancy M, Meyer FD, Lanning SP, Blake NK, Talbert LE, Giroux MJ (2002) Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci USA 99(3):1724–1729. https://doi.org/10.1073/pnas.022635299
Smidansky ED, Meyer FD, Blakeslee B, Weglarz TE, Greene TW, Giroux MJ (2007) Expression of a modified ADP-glucose pyrophosphorylase large subunit in wheat seeds stimulates photosynthesis and carbon metabolism. Planta 225(4):965–976. https://doi.org/10.1007/s00425-006-0400-3
Song Y, Zheng Q (2008) Influence of gliadin removal on strain hardening of hydrated wheat gluten during equibiaxial extensional deformation. J Cereal Sci 48(1):58–67. https://doi.org/10.1016/j.jcs.2007.08.001
Tanabe S (2005) A Novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell Online 17(3):776–790. https://doi.org/10.1105/tpc.104.024950
Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, Yokota T, Asami T, Kamakura T, Mori M (2009) BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151(2):669–680. https://doi.org/10.1104/pp.109.140806
Tang XJ, Peng C, Zhang J, Cai Y, You XM, Kong F, Yan HG, Wang GX, Wang L, Jin J, Chen WW, Chen XG, Ma J, Wang P, Jiang L, Zhang WW, Wan JM (2016) ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm. Plant Sci 249:70–83. https://doi.org/10.1016/j.plantsci.2016.05.010
Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C (2012) DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24(6):2562–2577. https://doi.org/10.1105/tpc.112.097394
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. https://doi.org/10.1093/bioinformatics/btp120
Tuncel A, Okita TW (2013) Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: expectations and unanticipated outcomes. Plant Sci 211:52–60. https://doi.org/10.1016/j.plantsci.2013.06.009
Wagner GP, Kin K, Lynch VJ (2012) Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131(4):281–285. https://doi.org/10.1007/s12064-012-0162-3
Wang ZY, Nakano T, Gendron J et al (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Development Cell 2:505–513
Wang L, Xu YY, Ma QB, Li D, Xu ZH, Chong K (2006) Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res 16(12):916–922. https://doi.org/10.1038/sj.cr.7310111
Wang L, Xu Y, Zhang C, Ma Q, Joo SH, Kim SK, Xu Z, Chong K (2008) OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3(10):e3521. https://doi.org/10.1371/journal.pone.0003521
Wang Z, Li W, Qi J, Shi P, Yin Y (2014) Starch accumulation, activities of key enzyme and gene expression in starch synthesis of wheat endosperm with different starch contents. J Food Sci Technol 51(3):419–429. https://doi.org/10.1007/s13197-011-0520-z
Wang W, Pan Q, Tian B, He F, Chen Y, Bai G, Akhunova A, Trick HN, Akhunov E (2019) Gene editing of the wheat homologs of TONNEAU1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J 100(2):251–264. https://doi.org/10.1111/tpj.14440
Wang J, Wang R, Mao X, Zhang J, Liu Y, Xie Q, Yang X, Chang X, Li C, Zhang X, Jing R (2020) RING finger ubiquitin E3 ligase gene TaSDIR1-4A contributes to grain size in common wheat. J Exp Bot. https://doi.org/10.1093/jxb/eraa271
Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, Matsumoto S, Fujioka S, Feldmann KA, Pennell RI (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20(8):2130–2145. https://doi.org/10.1105/tpc.107.055087
Xiong H, Guo H, Xie Y, Zhao L, Gu J, Zhao S, Li J, Liu L (2017) RNAseq analysis reveals pathways and candidate genes associated with salinity tolerance in a spaceflight-induced wheat mutant. Sci Rep 7(1):2731. https://doi.org/10.1038/s41598-017-03024-0
Yamamori M, Hirano H et al. (1993) Identification of three Wxproteinsin wheat. Biochemica Genetics 31:75-86
Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X (2010) Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci USA 107(13):6100–6105. https://doi.org/10.1073/pnas.0912333107
Yin Y, Wang ZY, Mora-Garcia S et al (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109:181–191
Yu K, Liu D, Chen Y, Wang D, Yang W, Yang W, Yin L, Zhang C, Zhao S, Sun J, Liu C, Zhang A (2019) Unraveling the genetic architecture of grain size in einkorn wheat through linkage and homology mapping and transcriptomic profiling. J Exp Bot 70(18):4671–4688. https://doi.org/10.1093/jxb/erz247
Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F, Xin P, Yan C, Chu J, Li HQ, Chen YQ (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31(9):848–852. https://doi.org/10.1038/nbt.2646
Zhang C, Bai MY, Chong K (2014) Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep 33(5):683–696. https://doi.org/10.1007/s00299-014-1578-7
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0100900), the Key Research and Development Program of Sichuan Province (2018NZDZX0002), Sichuan Science and Technology Program (2020YFH0140), the International Science & Technology Cooperation Project of Chengdu, Sichuan Province, China (2019-GH02-00078-Hz), and the Technological Innovation Project of Chengdu, Sichuan Province, China (2018-YF05-00059-SN).
Author information
Authors and Affiliations
Contributions
XJZ carried out analysis of RNA sequencing, qRT-PCR and wrote this article. NL and JJD analyzed the bioinformatics. QY carried out material preparation. JYL and HPT contribute to the material planting, the succession of mutant material and management. QTJ conceived and designed the experiments. PFQ, MD, JM, JRW, GYC, XJL, YMW, and YLZ partially participated in its design and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Supplementary Information
Figure S1. Hierarchical clustering heatmap of Pearson’s correlation coefficients between samples. The biological replicates of each sample are indicated as “1, 2.” The color scale represents the values of Pearson’s correlation coefficients.
Figure S2. GO terms of all DEGs of the SM482gs mutant compared with WT in different grain developmental stages. (A) GO terms of all upregulated DEGs in different comparisons. (B) GO terms of all downregulated DEGs in different comparisons. W: WT, M: SM482gs mutant. p≤0.05.
Figure S3. KEGG pathway enrichment of DEGs of the SM482gs mutant compared with WT in different grain developmental stages. W: WT, M: SM482gs mutant. p≤0.05.
Table S1. Top ten biological processes (BP) enriched among the down- and upregulated DEGs of the SM482gs mutant as compared with WT in different grain developmental stages. W: WT, M: SM482gs mutant.
Table S2. Top ten KEGG pathways enriched among the down- and upregulated DEGs of the SM482gs mutant as compared with WT in different grain developmental stages. W: WT, M: SM482gs mutant.
Table S3. Functional classification of DEGs in each k-means cluster. The numbers represent the numbers of genes.
Table S4. NCBI and transcript IDs of starch synthetase genes, BR-related homologous genes from Triticum species, rice, Arabidopsis, and storage protein genes.
Table S5. The detailed information and primers of genes analyzed for RT-qPCR.
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhong, X., Lin, N., Ding, J. et al. Genome-wide transcriptome profiling indicates the putative mechanism underlying enhanced grain size in a wheat mutant. 3 Biotech 11, 54 (2021). https://doi.org/10.1007/s13205-020-02579-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02579-6