Genetic Structure in Populations of Euterpe precatoria Mart. in the Brazilian Amazon
- 1Universidade Federal do Amazonas, Manaus, Brazil
- 2Centro Universitário UNIVEL, Avenida Tito Muffato, Cascavel, Brazil
- 3Faculdade de Ciências Agrícolas, Universidade Federal do Amazonas, Manaus, Brazil
- 4Embrapa Florestas, Colombo, Brazil
- 5Embrapa Amazônia Ocidental, Manaus, Brazil
- 6Luiz de Queiroz College of Agriculture, University of São Paulo, Piracicaba, Brazil
- 7Departamento de Biologia Vegetal, Instituto de Biologia, Universidade Estadual de Campinas (UNICAMP), Campinas, Brazil
- 8Escola Superior de Agricultura Luiz de Queiroz/Universidade de São Paulo (ESALQ/USP), Departamento de Ciências Florestais, Piracicaba, Brazil
Euterpe precatoria is a palm tree belonging to the Arecaceae family, occurring in Western and Central Brazilian Amazonia. Its fruit, which is very appreciated in the Amazon region, produces pulp that is consumed in fresh form. Its production is carried out almost exclusively by extractive farmers. In order to establish adequate strategies to sustain this genetic resource, we need knowledge about the diversity and genetic structure in natural populations. This study aimed to evaluate the influence of geographic distance on genetic structure in the main extractive populations of E. precatoria in the Brazilian Amazon. Leaves from 377 plants were collected in 19 populations located in 16 municipalities in the State of Amazonas and three in the State of Rondônia. Twelve microsatellite loci were used to genotype the plants. The diversity and genetic structure among populations were estimated. The average number of alleles per locus was 5.97. The observed heterozygosity means (HO) were higher than expected (HE) at the population level (HO = 0.72, HE = 0.66) and fixation index (f = -0.100) was negative. The FST value (0.1820) and the AMOVA results (Φ = 0.1796) showed population structure. The populations were clustered into three groups (K = 3) in the Bayesian analysis. The Discriminant Analysis of Principal Components (DAPC) confirmed eight clusters, with the populations close to those identified by the Bayesian analysis. The geographic differentiation was confirmed by the groupings obtained in the Structure analysis and the DACP function. Information related to phenotypic, genetic and environmental characterization of populations is important to guide conservation and management strategies and the formulation of public species management policies in Amazonia.
Introduction
Palm trees compose a group of plants of particular economic importance, especially in rural communities in tropical forests. More specifically, extractives provide building materials, fabrics, fuels, food, ornamental plants and medicines (Sosnowska and Balslev, 2009). These present greater abundance in regions closer to the Equator (Eiserhardt et al., 2011), but they are also found in other tropical and subtropical regions, with spatial patterns according to geographic variation. The Arecaceae family has more than 2,400 species grouped within the subfamilies Arecoideae, Calamoideae, Ceroxyloideae, Coryphoideae, and Nypoideae (Dransfield et al., 2005; Govaerts and Dransfield, 2005). In the Brazilian Amazon, the subfamily Arecoideae, tribe Euterpeae (Dransfield et al., 2005), has two palm trees of significant economic and social importance, Euterpe oleracea Mart. and Euterpe precatoria Mart (Kahn, 1991). The fruit of these species are exploited mainly by extractive agriculture. The production by extractivist farmers, of these two species, has allowed the Amazon region to be the largest producer of açaí in the national and international markets.
The açaí fruit is obtained from E. oleracea in Pará, Maranhão and Amapá states, while in the states of Amazonas, Acre and Rondônia it is extracted from E. precatoria. Between the years 1986 and 2010, the production of commercialized açaí fruit averaged 117,063.59 tons per year. This average production increased by 80.3% (211,100.63 tons) from 2011 to 2018. The state of Pará remains the largest producer of açaí fruit (58.4%), followed by the states of Amazonas (31.0%) and Maranhão (7.1%) (Instituto Brasileiro de Geografia e Estatística – IBGE, 2019). In the state of Amazonas, the fruit production increased more than 6,317.7%, from an average production of 1,013.17–65,021.8 tons per year in the last 8 years, by extractive exploitation from E. precatoria. This highly significant increase in production in recent years could be affecting the genetic variability of these species, leading to important impacts on the structure and genetic composition of populations due to anthropic interference. This calls for the development of public policies that can contribute to more sustainable management and development models for palm species to meet current demands without compromising the ability to serve future generations (García-Fernández et al., 2008).
We asked if current extraction of E. precatoria fruits would create a problem in the future for obtaining successful regenerants to replace the current parents. The sustainability of a fruit production system, for example E. precatoria, depends not only on the intensity of the fruit harvest, but also on other aspects of the process, such as the period of abundance and distribution of the resource, the strategy of growth and regeneration of the species, as well as other management practices associated with the harvest (Vallejo et al., 2014). Lack of natural regeneration of E. precatoria has already been identified in communities in Bolivia where palm heart production and exploitation were carried out in an extractive manner by cutting of the plant (Mostacedo and Fredericksen, 1999). Plant cutting is a standard practice in the economic exploitation of palm trees, as it is fast and easy (Bernal et al., 2011), but it is not a method that comports with conservation. Therefore, we need a better model for exploiting palm products, such as fruits, seeds, and other parts of the plant.
The success of natural regeneration will help guarantee the sustainability of managed forests in the long term (Mostacedo and Fredericksen, 1999). At the same time, ex situ conservation programs based on the establishment of active germplasm banks and E. precatoria genetic improvement programs are recommended to guarantee the long-term use of the species. Such programs will also result in genotypes with more diverse characteristics for farmers, such as higher fruit production per plant, less harvesting of individuals, and early production. No such ex situ conservation and genetic improvement programs are currently in practice in Brazil for E. precatoria. The current conservation strategy adopted is in situ, and it occurs empirically in an agroforestry production system (Noda, 2012), or within secondary forests.
E. precatoria is known locally as açaí-do-Amazonas, açaí-solitary, or açaí-de-terra-firma (Bovi and Castro, 1993) by its unique characteristics (FAO – Food and Agriculture Organization of the United Nations, 1987; Kang et al., 2012). Its occurrence is limited to the western and central Brazilian Amazon, but the species also occurs in the Amazonian borders of Peru, Brazil, Colombia (Kahn, 1991), and Bolivia (FAO – Food and Agriculture Organization of the United Nations, 1987; Bussmann and Zambrana, 2012). In addition to its use in the production of açaí, which has antioxidant and anti-inflammatory properties superior to those of E. oleracea (Kang et al., 2012), the species is also used in the construction of houses. Its roots are the source of medicinal preparations to treat malaria, hepatitis and other diseases (Bussmann and Zambrana, 2012). Another important characteristic is its evolutionary adaptation. Plants of this species grow in the forest (sub-canopy) in environments with both favorable and unfavorable soils, frequently forming small groups of plants, between 10 and 20 individuals (FAO – Food and Agriculture Organization of the United Nations, 1987), or among 50–250 plants/ha (Kahn, 1988). It also occurs in lower densities, around 2–3 trees.ha–1 (FAO – Food and Agriculture Organization of the United Nations, 1987), in places with rainfall in the range of 1,900–4,000 mm per year and average annual temperatures of 26°C (FAO – Food and Agriculture Organization of the United Nations, 1987). A study carried out on samples distributed throughout the Amazon, shows that E. precatoria is a common species and with Hyperdominant Patterns, in the Amazon Forest Types, with a maximum abundance of 168/ha and an average presence of 32.7% in all sampled areas (Steege et al., 2013). It is also of secondary successional class (Condé and Tonini, 2013).
Euterpe precatoria is a highly promising species; therefore, efforts are needed to establish strategies for domestication, conservation, and management of its natural populations with the support of riverside communities and farmers in the state of Amazonas in Brazil. This calls for research in the areas of management and conservation (in situ, ex situ, or on farm) in the short term to meet the high product demand, as well as stimulate the implementation of areas of monoculture of the species with more quality genetics. Accordingly, it will be necessary to monitor the diversity and genetic structure of natural E. precatoria populations, especially those exploited in an extractive manner, to conserve this species based on managing genetic viability for the long term, but without affecting the mechanisms of pollen and seed dispersal, or the spatial distribution of individuals and effective population size (Novello et al., 2018).
Therefore, we aimed to evaluate the patterns of diversity and genetic structure of the Amazonian E. precatoria palm tree in the main areas of its extractive production. It is expected that our results will become the basis for programs highlighting rational management and genetic conservation to foster continued economic sustainability.
Materials and Methods
Material
This study was carried out in the Amazon rainforest in two of the main producing states of E. precatoria. The plant material (leaves) of E. precatoria was collected in 19 municipalities (Figure 1), 16 in the state of Amazonas and three in the state of Rondônia. The municipalities are located on the margins of different hydrographic basins, which, for the most part, serve as the only route of river transport of people, food, and genetic material to other areas of the Amazon, mainly the capitals of these states. These municipalities are all within the geographic distribution of the species (Lorenzi, 2010) and present a substantial record of açaí in the aggregated data of the Brazilian Institute of Geography and Statistics (Instituto Brasileiro de Geografia e Estatística – IBGE, 2019). Each sampled collection site was called a population. Samples ranging from 17 to 20 plants were collected in each of the 19 populations, totaling 377 samples (Supplementary Table 1). The collection was carried out within the rules of SisGen (National System for the Management of Genetic Heritage and Associated Traditional Knowledge, Decree no 8.772, of May 11, 2016, which regulates Law no 13.123, of May 20, 2015, registration AA9537B).
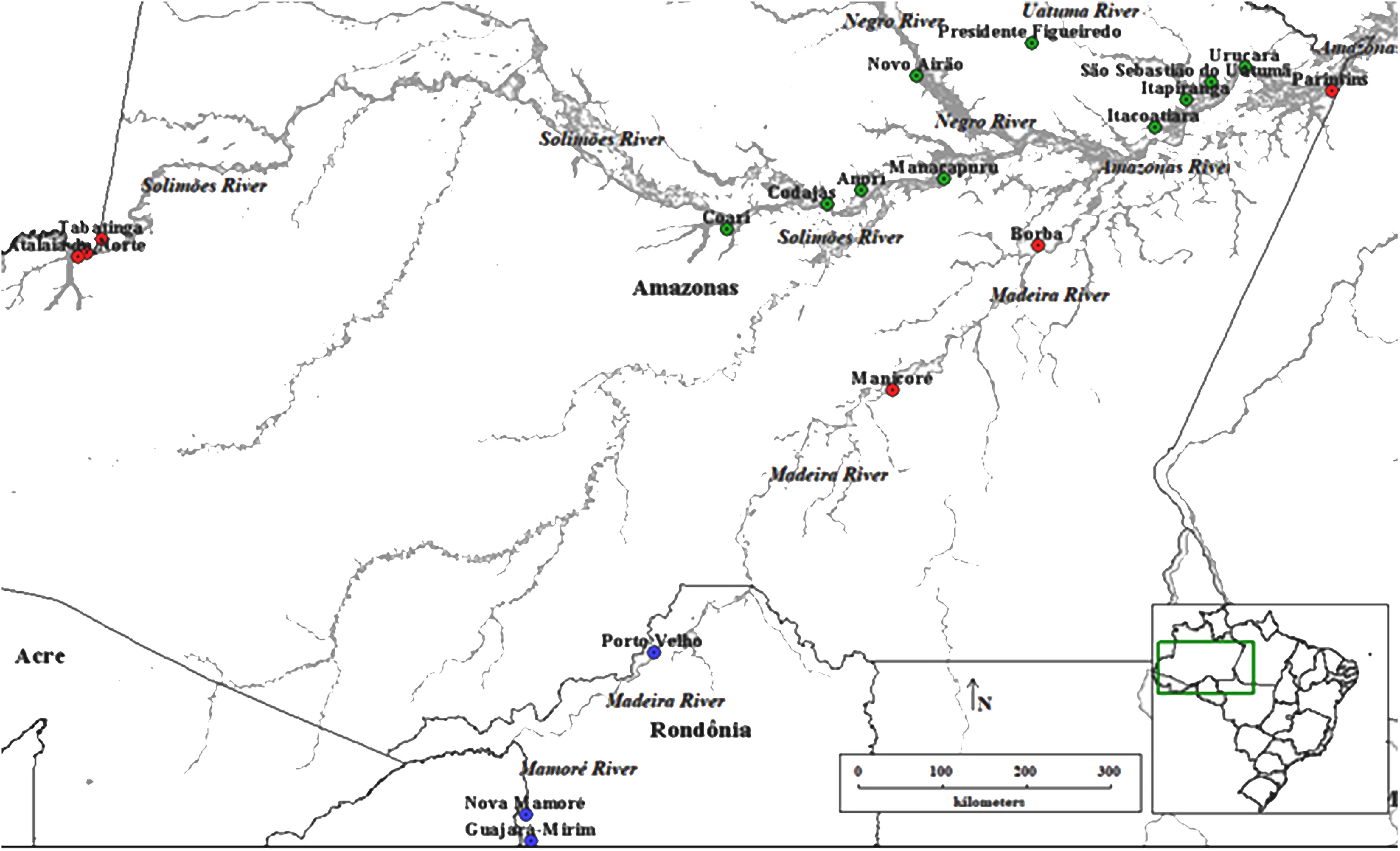
Figure 1. Collection of Euterpe precatoria vegetal materials in the areas of açaí-do-Amazonas extraction by traditional extractivist farmers in the states of Amazonas and Rondônia. Using the Structure software (Circle), three main groups formed by the 19 populations of E. precatoria according to the three main river basins, using the structure. Through the Discriminant Principal Component Analysis—DACP (square) eight groups formed by the proximity of the collection sites.
DNA Extraction and Genotyping
DNA extraction was performed using the CTAB protocol, as described by Doyle and Doyle (1990), and quantified with GelRed dye. Subsequently, the genomic DNA was standardized at 10 ng/μl for use in amplification. The 377 DNA samples were amplified by polymerase chain reaction (PCR), using 12 microsatellite loci developed for E. precatoria (Epr01, Epr02, Epr05, Epr13, Epr14, Epr15, Epr18, Epr19, Epr21, Epr31, Epr32, and Epr36) (Ramos et al., 2016). The amplification products were quantified with GelRed dye. The amplified PCR products were subjected to an automatic DNA analyzer by ABI 3130XL capillary electrophoresis in the Genetic Analyzer (Applied Biosystems). GeneScanTM -500 ROX® standard size (Life Technologies do Brasil Ltda.) was used to determine the size of the alleles. The amplified fragments were observed and analyzed with GeneMapper v4.0 software (Applied Biosystems).
Statistical Analysis
The genetic diversity, compared among the populations collected in the states of Amazonas and Rondônia, was obtained using the following genetic parameters: total number of alleles (AT), average number of alleles/locus (A), observed heterozygosity (HO) and expected heterozygosity (HE), fixation index (f), and Hardy-Weinberg equilibrium (HWE). These parameters were calculated using the divBasic function of the diveRsity package (Keenan et al., 2013) on the R platform (R Core Team, 2019).
The linkage disequilibrium (LD) and the number of private alleles (Ap) were calculated using the softwares GDA (Lewis and Zaykin, 2002), and the tests LD and Nm_private function of the genepop package (Rousset, 2008) on the R platform (R Core Team, 2019). HWE and LD were performed by Fisher’s exact test with 20,000 permutations. The significance level (P ≥ 0.05) of HWE and LD was adjusted with Bonferroni’s correction (Rice, 1989). The number of null alleles was calculated using the null function of the genepop package (Rousset, 2008).
For analysis of the genetic structure, Wright’s F statistics were used (Wright, 1951), including FIT (total inbreeding levels in individuals of all populations), FIS (fixation index within subpopulations owing to the reproductive system), and FST (genetic differentiation owing to subdivision), and calculations were performed using the algorithms of Weir and Cockerham (1984). An FST matrix was estimated from the general fixation index and in pairs of populations (Weir and Cockerham, 1984). The Wright F-statistics (1951) and the FST matrix were calculated and evaluated with significance (P ≤ 0.05) from 20,000 bootstrapping, using the diffCalc function of the diveRsity package (Keenan et al., 2013).
To verify the degree of genetic variation according to hierarchical levels among and within populations and among all individuals sampled, molecular variance analysis (AMOVA) was performed, followed by estimating the extent of genetic differentiation with phi-statistics, both using the poppr.amova function in the poppr package (Kamvar et al., 2014). The significance of the components of variance through the Monte-Carlo test, ΦCP, differentiation among populations, ΦCAP, differentiation among samples within populations, ΦCA, and differentiation among samples were evaluated using a permutation test implemented in the randtest function in the ape4 package (Chessel et al., 2004; Dray and Dufour, 2007; Dray et al., 2007; Bougeard and Dray, 2018) with 9,999 permutations.
In order to determine the correlation coefficient among the FST geographic and genetic distance matrices (Weir and Cockerham, 1984), the Mantel test (Mantel, 1967; Smouse et al., 1986) was performed with 999 replicates, using the mantel. rtest function of the ade4 package (Chessel et al., 2004; Dray and Dufour, 2007; Dray et al., 2007; Bougeard and Dray, 2018). The geographic distance matrix was calculated using the DIVA-GIS v7.5 program (DIVA-GIS, 2012).
Within the analysis of genetic structure, two Bayesian analysis techniques were performed to determine the number of clusters within the set of evaluated samples. In the first analysis, Structure software (Pritchard et al., 2000) from the Admixture model was used. This is normally applied to real or natural populations. The number of clusters (K) was defined from 1 to 21, and for each K, 10 iterations were performed with a Burn-in of 5,00,000 followed by 10,00,000 Markov Chain Monte Carlo (MCMC) iterations.
The number of clusters was estimated using the Evanno ΔK method, indicating that the most likely K is where the change is greatest in the second order rate of change in Pr (X| K) among successive K values (Evanno et al., 2005). With the K value selected, a consensus was reached on the iterations carried out in this cluster through CLUMPP v.1.1.2 (Cluster Matching program and Permutation Program) (Jakobsson and Rosenberg, 2007) and with Distruct v. 1.1 (Rosenberg, 2004), a graphical visualization of the population structure was performed. In the second analysis of the population genetic structure, Discriminant Principal Component Analysis (DACP) was used (Jombart et al., 2010), considering the clusters as unknown (Roullier et al., 2013). First, the probability of adherence of each sample to the different groups was determined, using k-means (Jombart et al., 2010). Then, the possible ideal number of clusters was identified, using k-means, comparing and discriminating possible groupings using Bayesian Information Criterion (BIC) (Jombart et al., 2010), using 10,00,000 permutations within the find.structure and dapc functions of the adegenet package 2.1.0 (Jombart and Ahmed, 2011) on the R platform (R Core Team, 2019).
Results
Genetic Diversity Indices
From the 12 loci analyzed, 11 were in Hardy-Weinberg equilibrium (P > 0.00417) (Supplementary Table 2), and the analysis of loci pairs showed 15.7% with linkage disequilibrium. The 12 highly polymorphic loci had a high content of genetic information, with a total of 211 alleles, ranging from 8 (Epr15) to 27 (Epr22) alleles per locus, with an average of 17.6 alleles per locus. The expected heterozygosity (HE) varied between 0.53 for Epr32 and 0.77 for Epr01 loci. The observed heterozygosities (HO) showed higher values than HE, with an exception for Epr02 (0.73) and Epr14 (0.66) loci. HO ranged between 0.56 for Epr32 locus and 0.86 for Epr13 and Eper05 loci. The average of all loci for HO was 0.72, higher than the average for HE (0.66). The fixation index (f) showed values below zero, which shows an excess of heterozygotes, although the Epr02 (f = 0.084) and Epr14 (f = 0.100) loci showed a small degree of inbreeding for the evaluated populations. However, the negative value found for the average (f = –0.108) shows the absence of inbreeding and excess heterozygotes for all populations and loci.
The 19 natural populations of E. precatoria showed high levels of genetic diversity. An average of 5.97 alleles per locus, ranging from 4.08 in Parintins to 7.92 for Guajará Mirim and Porto Velho populations (Table 1). High values were observed for HO and HE for all populations, with HO showing values equal to, or slightly higher than HE in 18 populations, with a single exception for the population of Atalaia do Norte, AM. The mean HO of all populations (0.72) was also higher than the HE (0.66). The average of f values per locus was below zero (f = –0.100), showing an excess of heterozygotes (Table 1). The allelic richness was 5.13.
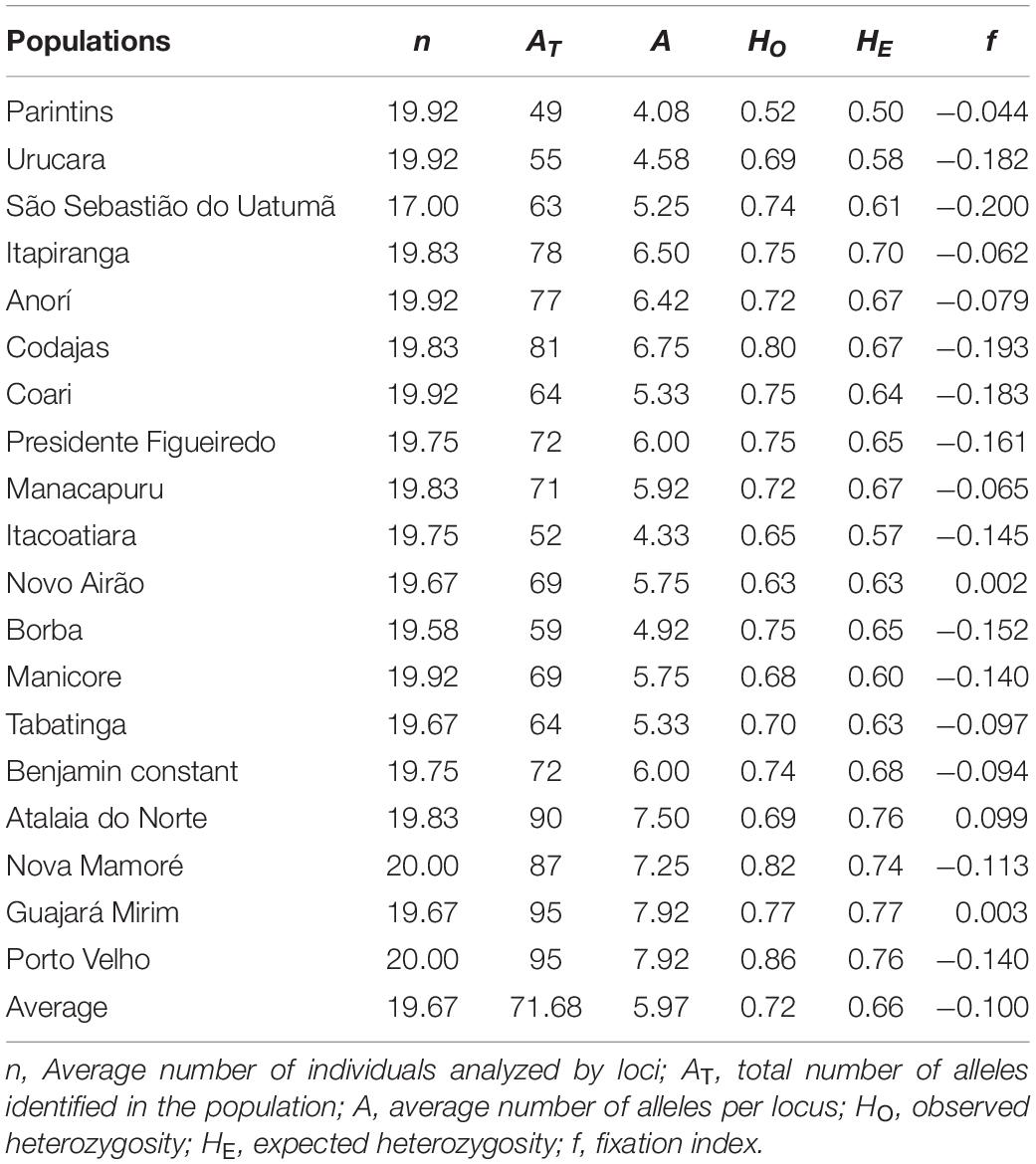
Table 1. Genetic diversity indices for 19 natural populations of E. precatoria analyzed with 12 microsatellite loci in the states of Amazonas and Rondônia.
The analyzes also showed a total of 37 private alleles observed in 16 populations, for all evaluated loci, except for the Epr36 locus (Supplementary Table 2), indicating a mean frequency of private alleles of 0.0986. Most of the studied loci showed linkage equilibrium, with percentages of LD that varied between zero percentage for the Borba population to 51.5% for the Atalaia do Norte population (Supplementary Table 2). Possible null alleles were detected for all microsatellite loci used. However, the frequency of these possible null alleles was found for some populations with frequencies less than 0.20. Null alleles with a frequency of less than 0.20 do not compromise population assessments (Dakin and Avise, 2004). Loci Epr22, Epr13, Epr14, and Epr19 presented higher frequencies in a single population. These alleles were maintained in the current study, for keeping their frequencies below 0.20.
Genetic Structure
Estimates of Wright’s F (Wright, 1951) in the 19 populations of E. precatoria sampled suggest that diversity is more concentrated within populations than among populations; however, high levels of genetic differentiation are indicated by an FST of 0.1820 (Wright, 1978). The estimates of FIT (0.1215) and FIS (-0.0740) were lower compared to FST (Table 2). The FST and FIT fixation indexes were outside the upper and lower limits of the bootstrappping, which indicates that the estimates are significantly different from zero. However, the FIS was not significant because it is lower than zero, indicating that the frequency of homozygotes is less than expected under EHW, showing an absence of inbreeding crosses (Table 2). In the FST estimates pair by pair (Table 3), most of the sampled populations showed significant differences. The largest statistical difference was observed among the sampled populations of Parintins and Itacoariara (39.57%) and the smallest among the populations of Anorí and Codajás (1.68%), the latter showing no statistical difference (p < 0.05) (Table 3).

Table 2. Results of Wright’s F-statistics estimates (Wright, 1951) obtained for 19 populations of Euterpe precatoria using 12 specific microsatellite loci.
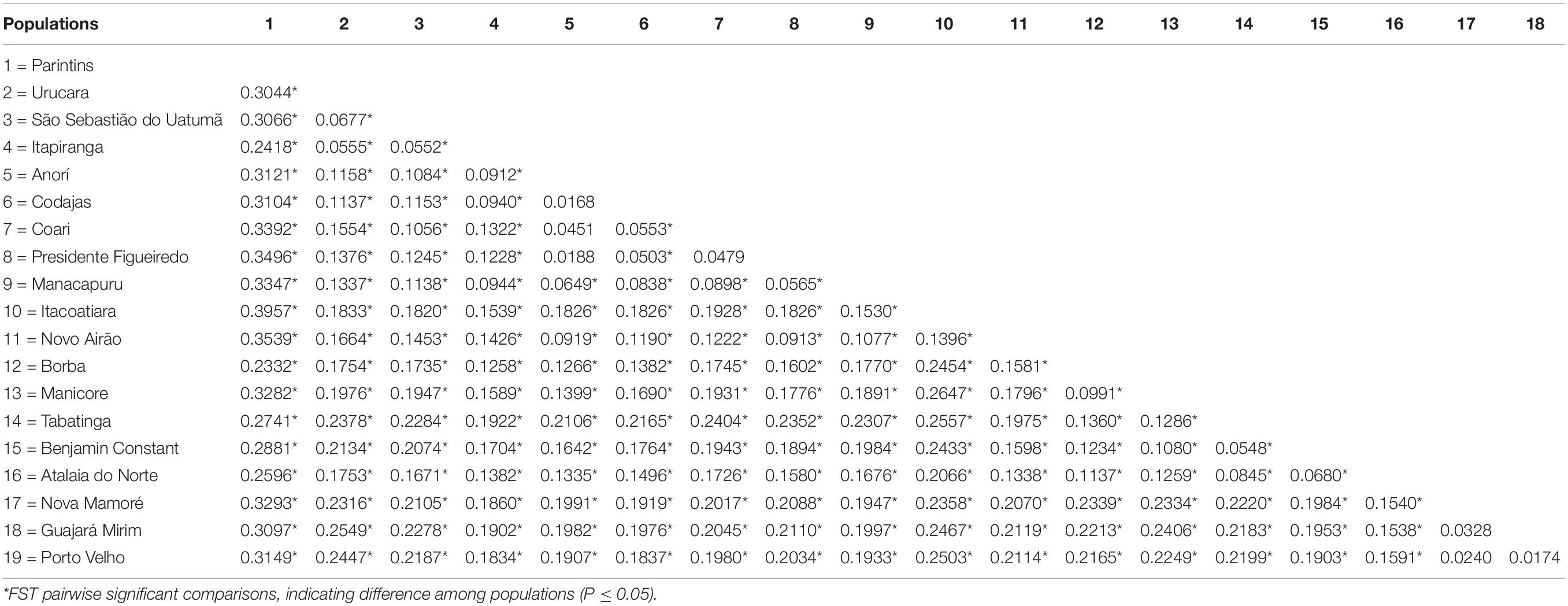
Table 3. Pairwise comparisons of the FST among the 19 populations of E. precatoria collected in the states of Amazonas and Rondonia.
The AMOVA results confirm the existence of genetic structure in the set of populations evaluated. Significant differences were observed among populations (18%; p = 0.004); however, most of the genetic variation occurred among the individuals evaluated (87.8%; p = 0.004) (Table 4). The significance of the components of variance of differentiation among populations (ΦCP = 0.1796), among samples within populations (ΦCAP = -0.0697) and among samples (ΦCA = 0.1224) is compatible with the results identified in statistical estimates of Wright’s F(1, 951), such as FST, FIS, and FIT, respectively. Under this scenario, the Mantel test, using the genetic matrices of FST and geographical distances of the populations, showed positive and significant correlations (r = 0.4044, p = 0.001), proving the existence of isolation by distance among the populations evaluated.
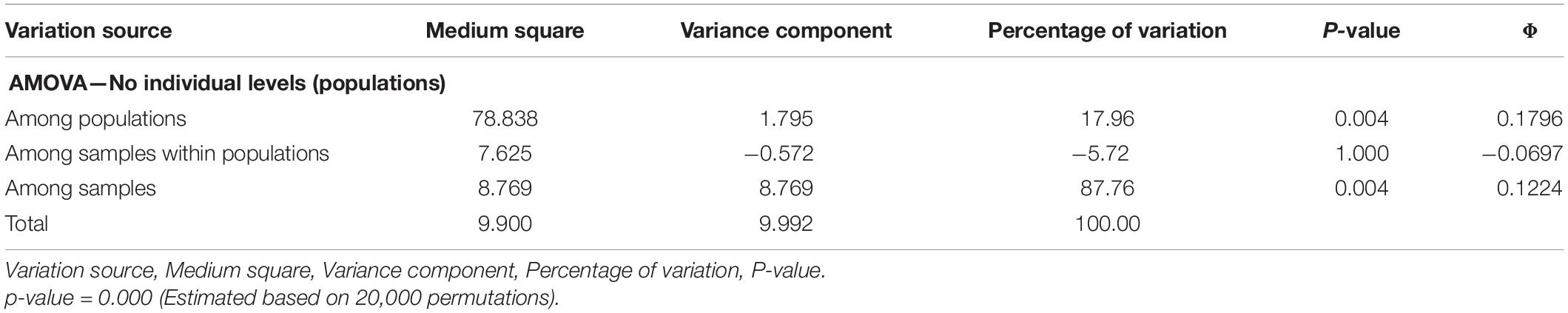
Table 4. Analysis of molecular variance (AMOVA) performed for 19 populations of E. precatoria collected in the states of Amazonas and Rondônia.
The estimate of the number of genetically homogeneous populations (K) among all individuals sampled in the 19 populations, as performed by the Structure software, showed the occurrence of three clusters (K = 3) (Figure 1). The classification of populations into three groups (K = 3) shows consistency in relation to geographical distance (Figure 3). The groups formed, considering K = 3, were as follows: group I covers the populations of the geographical basins of the Solimões, Negro, Uatumã and Amazonas Rivers (Ucurará, São Sebastião do Uatumã, Itapiranga, Anorí, Codajas, Coari, Presidente Figueiredo, Manacapuru, Itacoatiara, Novo Airão); group II is composed of the populations of the Solimões, Madeira and Amazonas Rivers (Parintins, Borba, Manicoré, Tabatinga, Benjamin Constant, Atalaia do Norte), and group III is found in populations of the Mamoré and Madeira Rivers (Nova Mamoré, Guajará Mirim and Porto Velho) (Figures 1, 2).
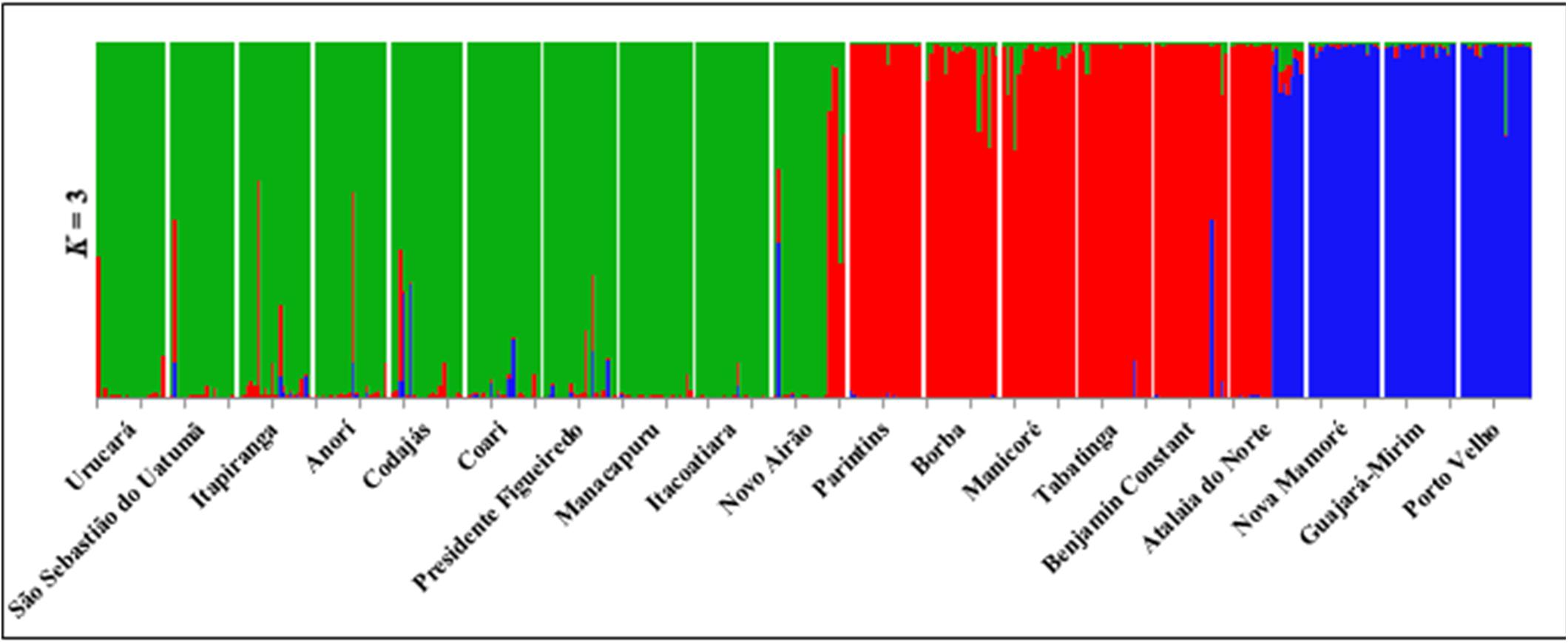
Figure 2. Structure of individuals from 19 populations of E. precatoria when grouped toward the North, South and Center of Amazonas.
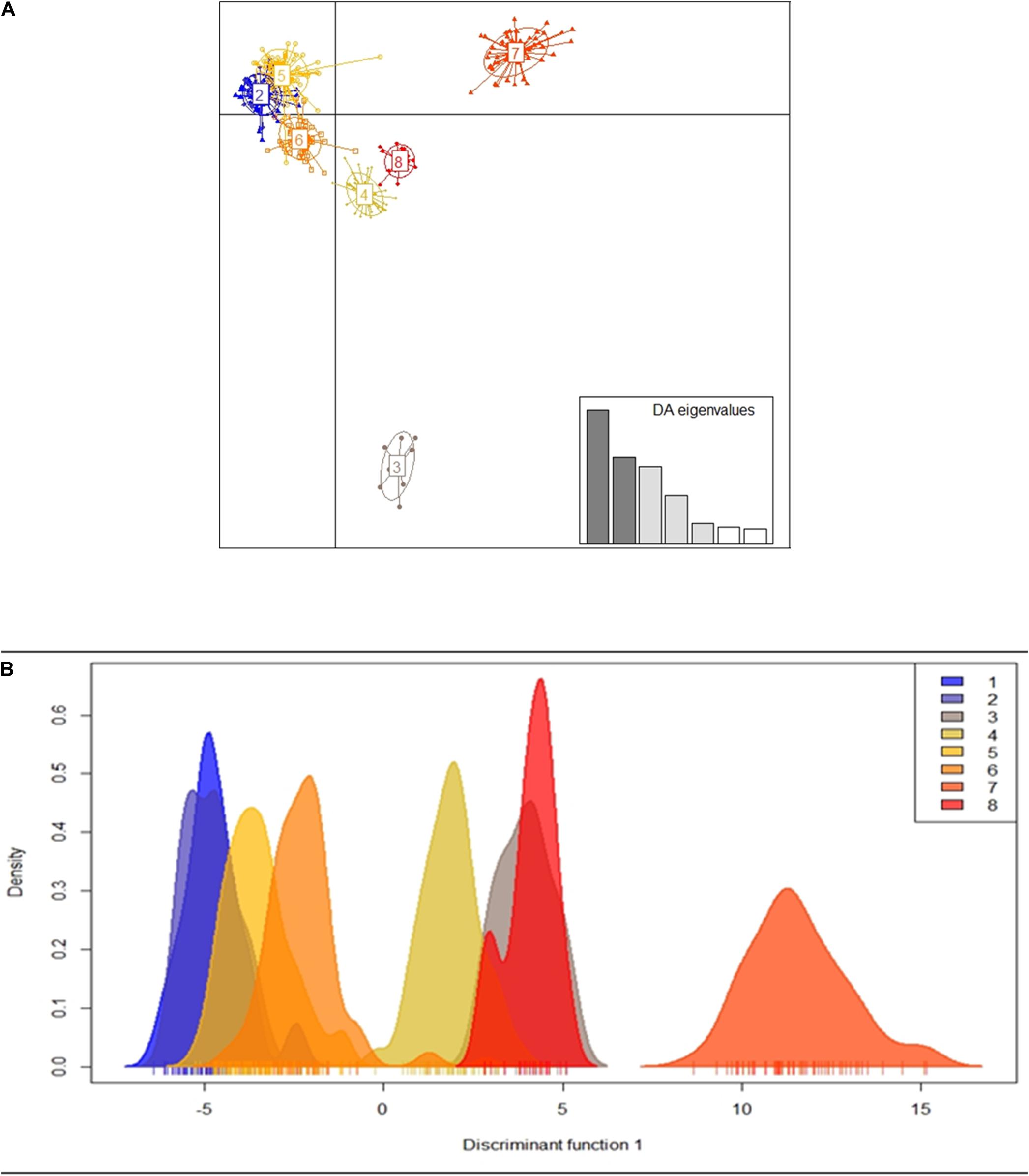
Figure 3. Dispersion graph of clusters formed by the k-means method (A) and density graph of individuals according to clusters identified along the discriminant function obtained in the DAPC for the 19 populations of E. precatoria (B).
Using DACP analysis, population structure is formed by eight groups (Figure 3 and Table 5). The seven eigenvalues obtained in the Discriminant Analysis (32.5, 21.1, 18.8, 11.7, 4.9, 4.1, and 3.5%) explain 96.7% of the genetic variation among populations, with the first three eigenvectors explaining a greater percentage of the variation (72.4%). The results obtained are different from those verified by the Bayesian analysis carried out in the Structure program. However, DACP groupings I, IV, V, VI, and VII are closely related to the hydrographic basins where the populations were sampled (Table 5). In DACP group I, the populations of Urucara and São Sebastiao do Uatumã are grouped within the Uatumã River. In group IV, the populations of Tabatinga, Benjamin Constant and Atalaia do Norte are grouped at the headwaters of the Solimões River. In group V, the populations of Anori, Codajas, and Manacapuru are at the center and end of the Solimões River basin. In group VI, Borba and Manicore are located on the Madeira River. In group VII, Nova Mamoré, Guajará Mirim and Porto Velho are part of the Mamoré River that originated the Madeira River.
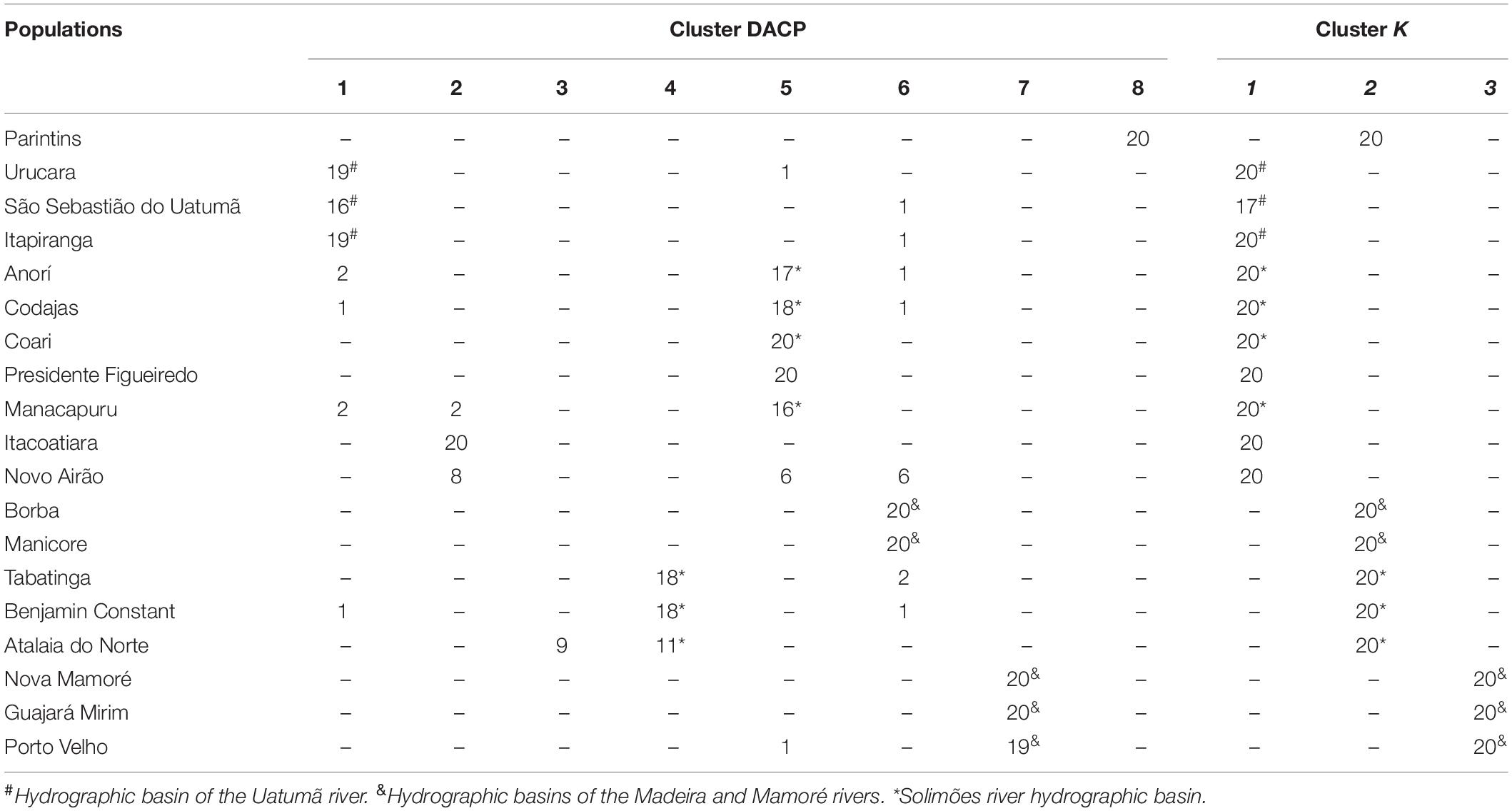
Table 5. Distribution of E. precatoria individuals among the groups formed by the Principal Component Discriminant Analysis (DACP) and Structure (K) based on 12 microsatellite loci.
Discussion
This is the first study to report on diversity and genetic structure of the natural populations of E. precatoria. The results will help define domestication and in situ and ex situ conservation strategies, as well as the management of natural or planted populations for the benefit of traditional Amazonian farmers and future enterprises. The E. precatoria plants sampled in the 19 populations under study revealed high levels of genetic diversity, mainly confirmed by the high content of genetic information observed in each index analyzed. The results on genetic diversity presented for this species are close to those of E. oleracea Mart. (Oliveira et al., 2010) and E. edulis (Novello et al., 2018). The negative values for the fixation indices (f) and the observed (HO) and expected (HE) heterozygosity coefficients indicate high genetic diversity (Nazareno and Reis, 2014), as confirmed by the excess of heterozygotes in the populations. The low f values and high HO also indicate that the result may be influenced by the reproductive system of the species (Ramos et al., 2011) which is allogamy (Ramos et al., 2018, 2019). Similar results were found in other palm species, such as Geonoma schottiana (Silva et al., 2011), Phoenix dactylifera (Arabnezhad et al., 2012) and Oenocarpus bataua (Ottewell et al., 2012), which could indicate a behavior common to plants of the family Arecaceae.
The high genetic diversity observed for the species is an important information for the domestication process of E. precatoria, even though the anthropic extraction may affect the genetic diversity of the species in the future, in the process of recruiting new plants, in the seedling bank. However, domestication generally begins with the exploration of wild plants, proceeds with the cultivation of plants selected in nature, not genetically different from wild plants, and ends with the fixation of morphological and genetic characteristics carried out by human selection (Pickersgill, 2007). Thus, the domestication process must be part of any proposals for genetic conservation and management in order to contribute adequately to the process of evolution and adaptation throughout the region of occurrence of the species and the planting zones (Kalinowski, 2004).
Traditionally, genetic analyses in natural populations may not present equilibrium conditions when based on the assumption of panmixia that is not always met (Oosterhout et al., 2006). We also observed a great geographical distance among many of the studied populations, and different evolutionary events may have taken place in each location, causing the absence of HWE for some of the initiators of certain populations.
The 19 populations of E. precatoria distributed in the Legal Amazon showed genetic differentiation when Wright’s F statistics (Hartl and Clark, 2010) was analyzed, indicating the existence of a population genetic structure according to the FST values. The results also confirm that a large part of the genetic variability observed for the set of populations sampled is mostly within populations, owing to the reproductive system (FIS). The populations also showed significant differences when analyzed in pairs (FST), also confirming the existence of genetic structure among the sampled populations (Wright, 1951). This result may be strongly influenced by the reproductive characteristics of the species, as well as the dispersion capacity of its genetic material, the degree of isolation of the population, the self-incompatibility system and allelic diversity that it presents (Leducq et al., 2011; Ramos et al., 2018).
The genetic structure observed also indicates that the sampled populations would be grouped according to the geographical distances within the hydrographic basins. Moreover, the occurrence of gene flow among populations is inversely proportional to the geographical distances among them. Isolation by distance has been previously identified to occur among populations of E. oleracea (Freeland et al., 2011). Thus, considering the geographical distance among populations of E. precatoria and the results of the comparisons of the FST analysis pair by pair, this result, hypothetically, shows that the gene flow that would be shared among these populations is proportional to the geographical distances among them because individuals are more likely to disperse to nearby locations (isolation by distance), confirming the population structure of the species. The positive and significant genetic correlation between geographical distance (as a dispersion limiter) and genetic distance supports this hypothesis and the dissimilarity in the composition of E. precatoria populations (Freeland et al., 2011; Kristiansen et al., 2012). Therefore, the allelic frequencies observed in the studied populations are related to geographic distances. This geographic differentiation information is confirmed by the groupings obtained in the analyses by the Structure program and the DACP function. The connectivity of the populations, according to the group they are in, is similar between both analyses (Figures 2, 3 and Table 5). For example, in the three municipalities of Nova Mamoré, Guajara Mirim and Porto Velho in the state of Rondonia, the same group was observed.
The results also suggest that the populations of E. precatoria would have started the process of formation of subpopulations in different parts of the Amazon, which could be related to the domestication process of E. precatoria, likely initiated by the Amerindians (Clement, 2001). This event may be closely related to their behavior because the Amerindians had a traditional subsistence system with high species diversification and building complex agroecosystems, including wood and non-wood products (Caviglia-Harris and Sills, 2005). Secondary dispersion is also carried out by birds and monkeys and also by man, transporting the fruit to consume, sell or plant in other areas. This seed dispersal will determine the colonization of new sites and migration among neighboring populations, especially if it is zoochoric because the range of seed dispersion can be substantially greater (Barluenga et al., 2011). However, the Acai-do-Amazonas, as well as the production of agricultural products already domesticated, incipiently domesticated, or collected in the forests by farmers, is traded among these local populations, and they mainly use river transport for this purpose. This is a dispersion vector that influences the movement of seeds, affording the plant with the capacity to persist, expand and colonize new habitats (Liu et al., 2013). This was also observed in the population of Presidente Figueiredo, which comes from fruits obtained by different government projects that were transported from Anori and Codajas.
It is believed that the genetic structure of populations may be related to the dispersion of seeds carried out among the extractive farmers themselves, who use river transport among the nearest communities. Therefore, sampling of seeds or propagules to compose tests of origins and progenies and/or commercial plantations, both for conservation and improvement, must consider the three groups identified in the analysis of genetic structure, as well as the estimated diversity among and within populations. This means that seed collections must be carried out across the three main hydrographic basins.
There is a concern about the intensification of fruit collection by extractive exploitation which is driven by the market (Instituto Brasileiro de Geografia e Estatística – IBGE, 2019) as it could be having significant impacts on genetic erosion in the near future, and, consequently, the reduction of fruit availability for frugivorous vertebrates, as well as the reduction of gene flow among populations (Ramos et al., 2018). To address this, restrictions on fruit harvesting should be carried out, such as avoiding fruit collection in years of low productivity and maintaining at least minus 50% of the fruits on mother palms for later consumption of fauna (Novello et al., 2018) and regeneration of new remnants.
In the process of ecological and dynamic succession of the forest with respect to the species E. precatoria, the plants of the populations may be showing high levels of genetic diversity at different ontogenetic ages (Ramos et al., 2018). The seeds are expected to represent a mixture of half sibs and full sibs (Bittencourt and Sebbenn, 2007). However, it is important to adopt a management plan that includes the destination of some E. precatoria fruits to the ecological community to maintain forest dynamics. Studies aimed at characterizing populations based on morpho-agronomic characters related to fruit production, as well as plant resistance and adaptation, will be important to identify the most productive individuals and progenies that can be used in different production systems, as well as enrich natural populations with the species. Information related to phenotypic, genetic and environmental characterization of populations is important to guide conservation and management strategies and the formulation of public species management policies in Amazonian watersheds.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, KT198662−KT198684.
Author Contributions
SR, RL, and ML contributed to all stages, by planning the study design, sampling the populations, laboratory analyses, statistical analysis of the data, and writing of the manuscript. EV, GD, AA-P, and JG contributed to the laboratory work. JM contributed to the sampling of the populations and contacts with farmers. AA and TF collaborated with writing of the manuscript. MW collaborated statistical analysis of the data. All authors read and revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, ET, declared a past co-authorship with several of the authors, GD, AA, EV, and AA-P to the handling editor.
Acknowledgments
We thank CNPq, FAPESP, CAPES, and FAPEAM for their financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.603448/full#supplementary-material
References
Arabnezhad, H., Bahar, M., Mohammadi, H. R., and Latifian, M. (2012). Development, characterization and use of microsatellite markers for germplasm analysis in date palm (Phoenix dactylifera L.). Sci. Hortic. 134, 150–156. doi: 10.1016/j.scienta.2011.11.032
Barluenga, M., Austerlitz, F., Elzinga, J. A., Teixeira, S., Goudet, J., and Bernasconi, G. (2011). Fine-scale spatial genetic structure and gene dispersal in Silene latifolia. Heredity 106, 13–24. doi: 10.1038/hdy.2010.38
Bernal, R., Torres, C., García, N., Isaza, C., Navarro, J., Vallejo, M. I., et al. (2011). Palm management in South America. Bot. Rev. 77, 607–646. doi: 10.1007/s12229-011-9088-6
Bittencourt, J. V. M., and Sebbenn, A. M. (2007). Patterns of pollen and seed dispersal in a small, fragmented population of the wind pollinated tree Araucaria angustifolia in southern Brazil. Heredity 99, 580–591. doi: 10.1038/sj.hdy.6801019
Bougeard, S., and Dray, S. (2018). Supervised multiblock analysis in R with the ade4 package. J. Stat. Softw. 86, 1–17. doi: 10.1007/978-1-4939-8850-1_1
Bovi, M. L. A., and Castro, A. (1993). “Assaí (Euterpe oleracea, palmae),” in Selected Species and Strategies to Enhance Income Generation from Amazonian Forests, eds J. W. Clay and C. R. Clement (Roma: Food and Agriculture Organization of the United Nations), 58–67.
Bussmann, R. W., and Zambrana, N. Y. P. (2012). Facing global markets-usage changes in Western Amazonian plants: the example of Euterpe precatoria Mart. and E. oleracea Mart. Acta Soc. Bot. Pol. 81, 257–261. doi: 10.5586/asbp.2012.032
Caviglia-Harris, J. L., and Sills, E. O. (2005). Land use and income diversification: comparing traditional and colonist populations in the Brazilian Amazon. Agric. Econ. 32, 221–237. doi: 10.1111/j.1574-0862.2005.00238.x
Chessel, D., Dufour, A., and Thioulouse, J. (2004). The ade4 package – I: one-table methods. R News 4, 5–10.
Clement, C. R. (2001). “Melhoramento de espécies nativas,” in Recursos Genéticos e Melhoramento – Plantas, eds L. L. Nass, A. C. C. Valois, I S. Melo, and M. C. Valadares-Inglis (Rondonópolis: Fundação de Apoio à Pesquisa Agropecuária de Mato Grosso), 423–441.
Condé, T. M., and Tonini, H. (2013). Fitossociologia de uma Floresta Ombrófila Densa na Amazônia Setentrional, Roraima, Brasil. Acta Amaz. 43, 247–260. doi: 10.1590/s0044-59672013000300002
Dakin, E. E., and Avise, J. C. (2004). Microsatellite null alleles in parentage analysis. Heredity 93, 504–509. doi: 10.1038/sj.hdy.6800545
DIVA-GIS (2012). User Manual, version 7.5. Available online at: http://www.diva-gis.org/docs/DIVA-GIS_manual_7.pdf (accessed June 01, 2013).
Dransfield, J., Uhl, N. W., Asmussen, C. B., Baker, W. J., Harley, M. M., and Lewis, C. E. (2005). A new phylogenetic classification of the palm family, Arecaceae. Kew Bull. 60, 559–569.
Dray, S., and Dufour, A. (2007). The ade4 Package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20. doi: 10.18637/jss.v022.i04
Dray, S., Dufour, A., and Chessel, D. (2007). The ade4 package – II: two-table and K-table methods. R News 7, 47–52.
Eiserhardt, W. L., Svenning, J. C., Kissling, W. D., and Balslev, H. (2011). Geographical ecology of the palms (Arecaceae): determinants of diversity and distributions across spatial scales. Ann. Bot. 108, 1391–1416. doi: 10.1093/aob/mcr146
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294x.2005.02553.x
FAO – Food and Agriculture Organization of the United Nations (1987). Especies Forestales Productoras de Frutas y Otros Alimentos, 3. Ejemplos de América Latina. Estudio FAO Montes 44/3. Roma: FAO, 265.
Freeland, J. R., Kirk, H., and Petersen, S. (2011). Molecular Ecology, 2nd Edn. Oxford: Wiley-Blackwell.
García-Fernández, C., Ruiz-Perez, M., and Wunder, S. (2008). Is multiple-use forest management widely implementable in the tropics? Forest Ecol. Manag. 256, 1468–1476. doi: 10.1016/j.foreco.2008.04.029
Hartl, D. L., and Clark, A. G. (2010). Princípios de Genética de Populações, 4th Edn. Porto Alegre: Artmed.
Instituto Brasileiro de Geografia e Estatística – IBGE (2019). Sistema IBGE de Recuperação Automática – SIDRA: Produção da Extração Vegetal e da Silvicultura, ano de 2017. Available online at: https://sidra.ibge.gov.br/tabela/289 (accessed October 29, 2019).
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Jombart, T., and Ahmed, I. (2011). Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. doi: 10.1093/bioinformatics/btr521
Jombart, T., Devillard, S., and Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. doi: 10.1186/1471-2156-11-94
Kahn, F. (1988). Ecology of economically important palms in Peruvian Amazonia. Adv. Econ. Bot. 6, 42–49.
Kahn, F. (1991). Palms as key swamp forest resources in Amazonia. Forest Ecol. Manag. 38, 133–142. doi: 10.1016/0378-1127(91)90139-m
Kalinowski, S. T. (2004). Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv. Genet. 5, 539–543. doi: 10.1023/b:coge.0000041021.91777.1a
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2:e281. doi: 10.7717/peerj.281
Kang, J., Thakali, K. M., Xie, C., Kondo, M., Tong, Y., Ou, B., et al. (2012). Bioactivities of açaí (Euterpe precatoria Mart.) fruit pulp, superior antioxidant and anti-inflammatory properties to Euterpe oleracea Mart. Food Chem. 133, 671–677. doi: 10.1016/j.foodchem.2012.01.048
Keenan, K., McGinnity, P., Cross, T. F., Crozier, W. W., and Prodohl, P. A. (2013). diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 4, 782–788. doi: 10.1111/2041-210x.12067
Kristiansen, T., Svenning, J. C., Eiserhardt, W. L., Pedersen, D., Brix, H., Kristiansen, S. M., et al. (2012). Environment versus dispersal in the assembly of western Amazonian palm communities. J. Biogeogr. 39, 1318–1332. doi: 10.1111/j.1365-2699.2012.02689.x
Leducq, J. B., Llaurens, V., Castric, V., Saumitou-Laprade, P., Hardy, O. J., and Vekemans, X. (2011). Effect of balancing selection on spatial genetic structure within populations: theoretical investigations on the self-incompatibility locus and empirical studies in Arabidopsis halleri. Heredity 106, 319–329. doi: 10.1038/hdy.2010.68
Lewis, P. O., and Zaykin, D. (2002). Genetic Data Analysis: Computer Programe for the Analyse of Allelic Data. Version 1.0.1. Available online at: https://phylogeny.uconn.edu/software/# (accessed November 21, 2020).
Liu, B., Su, J., Chen, J., Cui, G., and Ma, J. (2013). Anthropogenic halo disturbances alter landscape and plant richness: a ripple effect. PLoS One 8:e56109. doi: 10.1371/journal.pone.0056109
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220.
Mostacedo, B., and Fredericksen, T. S. (1999). Regeneration status of important tropical forest tree species in Bolivia: assessment and recommendations. Forest Ecol. Manag. 124, 263–273. doi: 10.1016/s0378-1127(99)00076-6
Nazareno, A. G., and Reis, M. S. D. (2014). At risk of population decline? an ecological and genetic approach to the threatened palm species Butia eriospatha (Arecaceae) of Southern Brazil. J. Hered. 105, 120–129. doi: 10.1093/jhered/est065
Noda, H. (2012). “In situ breeding and conservation of Amazonian horticultural species,” in Domestication and Breeding: Amazonian Species, eds A. Borém, M. T. G. Lopes, C. R. Clement, and H. Noda (Viçosa: Universidade Federal de Viçosa), 170–208.
Novello, M., Viana, J. P. G., Alves-Pereira, A., de Aguiar Silvestre, E., Nunes, H. F., Pinheiro, J. B., et al. (2018). Genetic conservation of a threatened Neotropical palm through community-management of fruits in agroforests and second-growth forests. Forest Ecol. Manag. 407, 200–209. doi: 10.1016/j.foreco.2017.06.059
Oliveira, M. S. P. D., Santos, J. B. D., Amorim, E. P., and Ferreira, D. F. (2010). Variabilidade genética entre acessos de açaizeiro utilizando marcadores microssatélites. Cienc. Agrotec. 34, 1253–1260. doi: 10.1590/s1413-70542010000500025
Oosterhout, C. V., Weetman, D., and Hutchinson, W. F. (2006). Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol. Ecol. Notes 6, 255–256. doi: 10.1111/j.1471-8286.2005.01082.x
Ottewell, K., Grey, E., Castillo, F., and Karubian, J. (2012). The pollen dispersal kernel and mating system of an insect-pollinated tropical palm, Oenocarpus bataua. Heredity 109, 332–339. doi: 10.1038/hdy.2012.40
Pickersgill, B. (2007). Domestication of plants in the Americas: insights from Mendelian and molecular genetics. Ann. Bot. 100, 925–940. doi: 10.1093/aob/mcm193
Pritchard, J. K., Stephens, M., and Donnelly, P. J. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
R Core Team (2019). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online at: http://www.R-project.org/ (accessed December 20, 2019).
Ramos, S. L. F., Dequigiovanni, G., Lopes, M. T. G., Veasey, E. A., De Macedo, J. L. V., Batista, J. S., et al. (2016). Microsatellite records for volume 8, issue 1: microsatellite markers for Euterpe precatoria Mart. (Arecaceae) a palm species used by extractive traditional farmers of Amazon. Conserv. Genet. Resour. 8, 43–81. doi: 10.1007/s12686-016-0522-2
Ramos, S. L. F., Dequigiovanni, G., Sebbenn, A. M., Lopes, M. T. G., Macêdo, J. L. V. D., Veasey, E. A., et al. (2018). Paternity analysis, pollen flow, and spatial genetic structure of a natural population of Euterpe precatoria in the Brazilian Amazon. Ecol. Evol. 8, 11143–11157. doi: 10.1002/ece3.4582
Ramos, S. L. F., Lopes, M. T. G., Lopes, R., Cunha, R. N. V. D., Macêdo, J. L. V. D., Contim, L. A. S., et al. (2011). Determination of the mating system of Tucumã palm using microsatellite markers. Crop Breed. Appl. Biot. 11, 181–185. doi: 10.1590/S1984-70332011000200011
Ramos, S. L. F., Lopes, M. T. G., Lopes, R., Dequigiovanni, G., Macedo, J. L. V. D., Sebbenn, A. M., et al. (2019). Mating system analysis of Açaí-do-Amazonas (Euterpe precatoria Mart.) using molecular markers. Crop Breed. Appl. Biot. 19, 126–130. doi: 10.1590/1984-70332019v19n1n17
Rice, W. R. (1989). Analyzing tables of statistical test. Evolution 43, 223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x
Rosenberg, N. A. (2004). Distruct: a program for the graphical display of population structure. Mol. Ecol. Notes 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
Roullier, C., Benoit, L., McKey, D. B., and Lebot, V. (2013). Historical collections reveal patterns of diffusion of sweet potato in Oceania obscured by modern plant movements and recombination. Proc. Natl. Acad. Sci. U.S.A. 110, 2205–2210. doi: 10.1073/pnas.1211049110
Rousset, F. (2008). genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Silva, M. S., Vieira, F. A., and Carvalho, D. (2011). Diversity and genetic structure in natural population of Geonoma schottian Mart (ARECACEAE): implications for conservation. Cerne 17, 195–201. doi: 10.1590/s0104-77602011000200006
Smouse, P. E., Long, J. C., and Sokal, R. R. (1986). Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 35, 627–632. doi: 10.2307/2413122
Sosnowska, J., and Balslev, H. (2009). American palm ethnomedicine: a meta-analysis. J. Ethnobiol. Ethnomed. 5:43.
Steege, H. T., Pitman, N. C., Sabatier, D., Baraloto, C., Salomão, R. P., Guevara, J. E., et al. (2013). Hyperdominance in the Amazonian tree Flora. Science 342:1243092. doi: 10.1126/science.1243092
Vallejo, M. I., Galeano, G., Bernal, R., and Zuidema, P. A. (2014). The fate of populations of Euterpe oleracea harvested for palm heart in Colombia. Forest Ecol. Manag. 318, 274–284.
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-stattistic for the analysis of population structure. Evolution 38, 1358–1370.
Keywords: Amazon, Arecaceae, microsatellites, açaí-do-Amazonas, genetic diversity, genetic structure
Citation: Ramos SLF, Dequigiovanni G, Lopes MTG, Aguiar AV, Lopes R, Veasey EA, Macêdo JLV, Alves-Pereira A, Fraxe TJP, Wrege MS and Garcia JN (2021) Genetic Structure in Populations of Euterpe precatoria Mart. in the Brazilian Amazon. Front. Ecol. Evol. 8:603448. doi: 10.3389/fevo.2020.603448
Received: 06 September 2020; Accepted: 10 December 2020;
Published: 11 January 2021.
Edited by:
Lawrence Hurd, Washington and Lee University, United StatesReviewed by:
Christina Vinson, University of Brasilia, BrazilEvandro Vagner Tambarussi, State University of Midwest Paraná, Brazil
Copyright © 2021 Ramos, Dequigiovanni, Lopes, Aguiar, Lopes, Veasey, Macêdo, Alves-Pereira, Fraxe, Wrege and Garcia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Teresa Gomes Lopes, mtglopes@hotmail.com
 Santiago Linorio Ferreyra Ramos1
Santiago Linorio Ferreyra Ramos1  Maria Teresa Gomes Lopes
Maria Teresa Gomes Lopes Ricardo Lopes
Ricardo Lopes Alessandro Alves-Pereira
Alessandro Alves-Pereira Marcos Silveira Wrege
Marcos Silveira Wrege