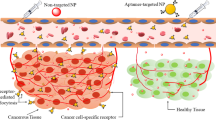
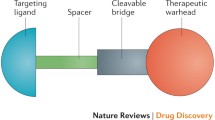
Abstract
Targeted treatment of cancer hinges on the identification of specific intracellular molecular receptors on cancer cells to stimulate apoptosis for eventually inhibiting growth; the development of novel ligands to target biomarkers expressed by the cancer cells; and the creation of novel multifunctional carrier systems for targeted delivery of anticancer drugs to specific malignant sites. There are numerous receptors, antigens, and biomarkers that have been discovered as oncological targets (oncotargets) for cancer diagnosis and treatment applications. Oncotargets are critically important to navigate active anticancer drug ingredients to specific disease sites with no/minimal effect on surrounding normal cells. In silico techniques relating to genomics, proteomics, and bioinformatics have catalyzed the discovery of oncotargets for various cancer types. Effective oncotargeting requires high-affinity probes engineered for specific binding of receptors associated with the malignancy. Computational methods such as structural modeling and molecular dynamic (MD) simulations offer opportunities to structurally design novel ligands and optimize binding affinity for specific oncotargets. This article proposes a streamlined approach for the development of ligand-oncotarget bioaffinity systems via integrated structural modeling and MD simulations, making use of proteomics, genomic, and X-ray crystallographic resources, to support targeted diagnosis and treatment of cancers and tumors.



Similar content being viewed by others
References
Callari, M., Gandellini, P., Skvortsova, I., & Span, P. N. (2018). Predicting and understanding cancer response to treatment. Disease Markers. https://doi.org/10.1155/2018/6159214.
Ferlay, J., Colombet, M., Soerjomataram, I., Mathers, C., Parkin, D. M., Piñeros, M., et al. (2019). Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. International Journal of Cancer, 144, 1941–1953.
Hinck, L., & Näthke, I. (2014). Changes in cell and tissue organization in cancer of the breast and colon. Current Opinion in Cell Biology, 26, 87–95.
Cooper, G. (2000). The cell: A molecular approach (2nd ed.). Sinauer Associates: Sunderland.
American Society of Cancer. (2015). Cancer facts and figures 2015. Atlanta: American Society of Cancer.
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68, 394–424.
Kim, R. (2017). Anesthetic technique and cancer recurrence in oncologic surgery: unraveling the puzzle. Cancer and Metastasis Reviews, 36, 159–177.
Liu, J., Yang, Y., Zhu, W., Yi, X., Dong, Z., Xu, X., et al. (2016). Nanoscale metal−organic frameworks for combined photodynamic & radiation therapy in cancer treatment. Biomaterials, 97, 1–9.
Van Driel, W. J., Koole, S. N., Sikorska, K., Schagen van Leeuwen, J. H., Schreuder, H. W. R., Hermans, R. H. M., et al. (2018). Hyperthermic intraperitoneal chemotherapy in ovarian cancer. New England Journal of Medicine, 378, 230–240.
Vertosick, E. A., Assel, M., Tokita, H. K., Zafirova, Z., Vickers, A. J., Simon, B. A., & Twersky, R. (2019). Suitability of outpatient or ambulatory extended recovery cancer surgeries for obese patients. Journal of Clinical Anesthesia, 58, 111–116.
Martin, O. A., Anderson, R. L., Narayan, K., & MacManus, M. P. (2017). Does the mobilization of circulating tumour cells during cancer therapy cause metastasis? Nature Reviews Clinical Oncology, 14, 32.
Morris, D., Tu, D., Tehfe, M. A., Nicholas, G. A., Goffin, J. R., Gregg, R. W., et al. (2016). A Randomized phase II study of reolysin in patients with previously treated advanced or metatstatic non small cell lung cancer (NSCLC) receiving standard salvage chemotherapy-canadian cancer trials group IND 211. Atlanda: American Society of Clinical Oncology.
Shamsi, M., Sedaghatkish, A., Dejam, M., Saghafian, M., Mohammadi, M., & Sanati-Nezhad, A. (2018). Magnetically assisted intraperitoneal drug delivery for cancer chemotherapy. Drug Delivery, 25, 846–861.
Lorusso, D., Bria, E., Costantini, A., Di Maio, M., Rosti, G., & Mancuso, A. (2017). Patients’ perception of chemotherapy side effects: Expectations, doctor–patient communication and impact on quality of life–An Italian survey. European Journal of Cancer Care, 26, e12618.
Yin, L., Lu, S., Zhu, J., Zhang, W., & Ke, G. (2019). Ovarian transposition before radiotherapy in cervical cancer patients: Functional outcome and the adequate dose constraint. Radiation Oncology, 14, 100.
Anghileri, L. J., & Robert, J. (2019). Hyperthermia in cancer treatment. Boca Raton: CRC Press.
He, Z., Liu, P., Zhang, S., Yan, J., Wang, M., Cai, Z., et al. (2019). A freezing-induced turn-on imaging modality for real-time monitoring of cancer cells in cryosurgery. Angewandte Chemie International Edition, 58, 3834–3837.
Qiu, W., Zhang, H., Chen, X., Song, L., Cui, W., Ren, S., et al. (2019). A glypican-1-targeted and gmcitabine-loaded biocompatible nanoplatform for pancreatic cancer near-infrared fluorescence/magnetic resonance multimodal imaging and therapy. Magnetic Resonance Multimodal Imaging and Therapy. https://doi.org/10.2139/ssrn.3328649.
Chang, D., Lim, M., Goos, J. A. C. M., Qiao, R., Ng, Y. Y., Mansfeld, F. M., et al. (2018). Biologically targeted magnetic hyperthermia: Potential and limitations. Frontiers in Pharmacology, 9, 831.
Mucciardi, G., Magno, C., Inferrera, A., & Lugnani, F. (2016). Cryosurgery and irreversible electroporation: The state of the art, advantages, and limitations. Handbook of Electroporation. https://doi.org/10.1007/978-3-319-32886-7_110.
Charmsaz, S., & Boyd, A. W. (2017). Eph receptors as oncotargets. Oncotarget, 8, 81727.
Jiao, Y., Wang, Y., Guo, S., & Wang, G. (2017). Glutathione peroxidases as oncotargets. Oncotarget, 8, 80093.
Chen, X., Zheng, Q., Li, W., Lu, Y., Ni, Y., Ma, L., & Fu, Y. (2018). SOX5 induces lung adenocarcinoma angiogenesis by inducing the expression of VEGF through STAT3 signaling. OncoTargets and Therapy, 11, 5733.
Chen, Y.-H., Lin, T.-T., Wu, Y.-P., Li, X.-D., Chen, S.-H., Xue, X.-Y., et al. (2019). Identification of key genes and pathways in seminoma by bioinformatics analysis. OncoTargets and Therapy, 12, 3683.
Wang, S., Yu, Z.-H., & Chai, K.-Q. (2019). Identification of EGFR as a novel key gene in clear cell renal cell carcinoma (ccRCC) through bioinformatics analysis and meta-analysis. BioMed Research International. https://doi.org/10.1155/2019/6480865.
American Cancer Society. (2019). Cancer facts and figures 2019. Atlanta: American Cancer Society.
Dela Cruz, C. S., Tanoue, L. T., & Matthay, R. A. (2011). Lung cancer: epidemiology, etiology, and prevention. Clinics in Chest Medicine, 32, 605–644.
Zhang, Q., Zeng, L., Chen, Y., Lian, G., Qian, C., Chen, S., et al. (2016). Pancreatic cancer epidemiology, detection, and management. Gastroenterology Research and Practice, 2016, 8962321.
Akram, M., Iqbal, M., Daniyal, M., & Khan, A. U. (2017). Awareness and current knowledge of breast cancer. Biological Research, 50, 33–33.
Hoffman, A. M., & Cairns, P. (2011). Epigenetics of kidney cancer and bladder cancer. Epigenomics, 3, 19–34.
De Minicis, S., Kisseleva, T., Francis, H., Baroni, G. S., Benedetti, A., Brenner, D., et al. (2013). Liver carcinogenesis: Rodent models of hepatocarcinoma and cholangiocarcinoma. Digestive and Liver Disease, 45, 450–459.
Cuzick, J., Thorat, M. A., Andriole, G., Brawley, O. W., Brown, P. H., Culig, Z., et al. (2014). Prevention and early detection of prostate cancer. The Lancet Oncology, 15, e484–e492.
Terwilliger, T., & Abdul-Hay, M. (2017). Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer Journal, 7, e577–e577.
Shanbhag, S., & Ambinder, R. F. (2018). Hodgkin lymphoma: A review and update on recent progress. CA: A Cancer Journal for Clinicians, 68, 116–132.
Nabors, L. B., Ammirati, M., Bierman, P. J., Brem, H., Butowski, N., Chamberlain, M. C., et al. (2013). Central nervous system cancers. Journal of the National Comprehensive Cancer Network, 11, 1114–1151.
Fisher, R., Pusztai, L., & Swanton, C. (2013). Cancer heterogeneity: Implications for targeted therapeutics. British Journal of Cancer, 108, 479–485.
Meacham, C. E., & Morrison, S. J. (2013). Tumour heterogeneity and cancer cell plasticity. Nature, 501, 328–337.
Joensuu, H., & Dimitrijevic, S. (2001). Tyrosine kinase inhibitor imatinib (STIS71) as an anticancer agent for solid tumours. Annals of Medicine, 33, 451–455.
Li, M., Chen, W.-D., Papadopoulos, N., Goodman, S. N., Bjerregaard, N. C., Laurberg, S., et al. (2009). Sensitive digital quantification of DNA methylation in clinical samples. Nature Biotechnology, 27, 858–863.
Doi, A., Park, I.-H., Wen, B., Murakami, P., Aryee, M. J., Irizarry, R., et al. (2009). Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature Genetics, 41, 1350–1353.
Esteller, M. (2007). Epigenetic gene silencing in cancer: The DNA hypermethylome. Human Molecular Genetics, 16, R50–R59.
Whitwell, H. J., Worthington, J., Blyuss, O., Gentry-Maharaj, A., Ryan, A., Gunu, R., et al. (2020). Improved early detection of ovarian cancer using longitudinal multimarker models. British Journal of Cancer. https://doi.org/10.1038/s41416-019-0718-9.
Eftimie, R., & Hassanein, E. (2018). Improving cancer detection through combinations of cancer and immune biomarkers: A modelling approach. Journal of Translational Medicine, 16, 73.
Afonso, J., Santos, L. L., Longatto-Filho, A., & Baltazar, F. (2020). Competitive glucose metabolism as a target to boost bladder cancer immunotherapy. Nature Reviews Urology. https://doi.org/10.1038/s41585-019-0263-6.
Padma, V. V. (2015). An overview of targeted cancer therapy. Biomedicine (Taipei), 5, 19–19.
Yap, T. A., & Workman, P. (2012). Exploiting the cancer genome: Strategies for the discovery and clinical development of targeted molecular therapeutics. Annual Review of Pharmacology and Toxicology, 52, 549–573.
Kwak, E. L., Bang, Y. J., Camidge, D. R., Shaw, A. T., Solomon, B., Maki, R. G., et al. (2010). Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. New England Journal of Medicine, 363, 1693–1703.
Chapman, P. B., Hauschild, A., Robert, C., Haanen, J. B., Ascierto, P., Larkin, J., et al. (2011). Improved survival with vemurafenib in melanoma with BRAF V600E mutation. New England Journal of Medicine, 364, 2507–2516.
Tian, X., Yan, L., Zhang, D., Guan, X., Dong, B., Zhao, M., & Hao, C. (2016). PTK7 overexpression in colorectal tumors: Clinicopathological correlation and prognosis relevance. Oncology Reports, 36, 1829–1836.
Kim, J. H., Kwon, J., Lee, H. W., Kang, M. C., Yoon, H. J., Lee, S. T., & Park, J. H. (2014). Protein tyrosine kinase 7 plays a tumor suppressor role by inhibiting ERK and AKT phosphorylation in lung cancer. Oncology Reports, 31, 2708–2712.
Ahn, T., Roberts, M. J., Abduljabar, A., Joshi, A., Perera, M., Rhee, H., et al. (2019). A review of prostate-specific membrane antigen (PSMA) positron emission tomography (PET) in renal cell carcinoma (RCC). Molecular Imaging and Biology, 21, 799–807.
Sun, H., Zhu, X., Lu, P. Y., Rosato, R. R., Tan, W., & Zu, Y. (2014). Oligonucleotide aptamers: New tools for targeted cancer therapy. Molecular Therapy: Nucleic Acids, 3, e182.
Aravind, A., Jeyamohan, P., Nair, R., Veeranarayanan, S., Nagaoka, Y., Yoshida, Y., et al. (2012). AS1411 aptamer tagged PLGA-lecithin-PEG nanoparticles for tumor cell targeting and drug delivery. Biotechnology and Bioengineering, 109, 2920–2931.
Zhang, J., Chen, R., Fang, X., Chen, F., Wang, Y., & Chen, M. (2015). Nucleolin targeting AS1411 aptamer modified pH-sensitive micelles for enhanced delivery and antitumor efficacy of paclitaxel. Nano Research, 8, 201–218.
Meng, Y., Xu, B.-Q., Fu, Z.-G., Wu, B., Xu, B., Chen, Z.-N., & Li, L. (2015). Cytoplasmic EpCAM over-expression is associated with favorable clinical outcomes in pancreatic cancer patients with Hepatitis B virus negative infection. International Journal of Clinical and Experimental Medicine, 8, 22204–22216.
Tayama, S., Motohara, T., Narantuya, D., Li, C., Fujimoto, K., Sakaguchi, I., et al. (2017). The impact of EpCAM expression on response to chemotherapy and clinical outcomes in patients with epithelial ovarian cancer. Oncotarget, 8, 44312–44325.
Fourcade, J., Sun, Z., Chauvin, J.-M., Ka, M., Davar, D., Pagliano, O., et al. (2018). CD226 opposes TIGIT to disrupt Tregs in melanoma. JCI Insight, 3, e121157.
Bottcher, J. P., Bonavita, E., Chakravarty, P., Blees, H., Cabeza-Cabrerizo, M., Sammicheli, S., et al. (2018). NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell, 172, 1022–1037.
Meng, X. Y., Zhang, H. X., Mezei, M., & Cui, M. (2011). Molecular docking: A powerful approach for structure-based drug discovery. Current Computer-Aided Drug Design, 7, 146–157.
Thomas, R. K., Baker, A. C., DeBiasi, R. M., Winckler, W., LaFramboise, T., Lin, W. M., et al. (2007). High-throughput oncogene mutation profiling in human cancer. Nature Genetics, 39, 347–351.
Ding, L., Ley, T. J., Larson, D. E., Miller, C. A., Koboldt, D. C., Welch, J. S., et al. (2012). Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature, 481, 506–510.
Gerlinger, M., Rowan, A. J., Horswell, S., Math, M., Larkin, J., Endesfelder, D., et al. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New England Journal of Medicine, 366, 883–892.
Vishwakarma, M., & Piddini, E. (2020). Outcompeting cancer. Nature Review Cancer. https://doi.org/10.1038/s41568-020-0261-2.
Baker, N. E. (2017). Mechanisms of cell competition emerging from Drosophila studies. Current Opinion in Cell Biology, 48, 40–46.
Di Gregorio, A., Bowling, S., & Rodriguez, T. A. (2016). Cell competition and its role in the regulation of cell fitness from development to cancer. Developmental Cell, 38, 621–634.
Claveria, C., & Torres, M. (2016). Cell competition: Mechanisms and physiological roles. Annual Review of Cell and Developmental Biology, 32, 411–439.
Wagstaff, L., Goschorska, M., Kozyrska, K., Duclos, G., Kucinski, I., Chessel, A., et al. (2016). Mechanical cell competition kills cells via induction of lethal p53 levels. Nature Communications, 7, 11373.
Levayer, R., Dupont, C., & Moreno, E. (2016). Tissue crowding induces caspase-dependent competition for space. Current Biology: CB, 26, 670–677.
Vincent, J. P., Kolahgar, G., Gagliardi, M., & Piddini, E. (2011). Steep differences in wingless signaling trigger Myc-independent competitive cell interactions. Developmental Cell, 21, 366–374.
Moreno, E., Basler, K., & Morata, G. (2002). Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature, 416, 755–759.
Rhiner, C., Lopez-Gay, J. M., Soldini, D., Casas-Tinto, S., Martin, F. A., Lombardia, L., & Moreno, E. (2010). Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila. Developmental Cell, 18, 985–998.
Madan, E., Pelham, C. J., Nagane, M., Parker, T. M., Canas-Marques, R., Fazio, K., et al. (2019). Flower isoforms promote competitive growth in cancer. Nature, 572, 260–264.
Ma, J., Wang, J., Ghoraie, L. S., Men, X., Liu, L., & Dai, P. (2019). Network-based method for drug target discovery at the isoform level. Scientific Reports, 9, 13868.
Maley, C. C., Aktipis, A., Graham, T. A., Sottoriva, A., Boddy, A. M., Janiszewska, M., et al. (2017). Classifying the evolutionary and ecological features of neoplasms. Nature Reviews Cancer, 17, 605–619.
Yang, B., Zhang, J., Yin, Y., & Zhang, Y. (2013). Network-based inference framework for identifying cancer genes from gene expression data. BioMed Research International. https://doi.org/10.1155/2013/401649.
Hori, S. S., Lutz, A. M., Paulmurugan, R., & Gambhir, S. S. (2017). A Model-based personalized cancer screening strategy for detecting early-stage tumors using blood-borne biomarkers. Cancer Research, 77, 2570–2584.
Hori, S. S., & Gambhir, S. S. (2011). Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Science Translational Medicine, 3, 109–116.
Kwong, G. A., Dudani, J. S., Carrodeguas, E., Mazumdar, E. V., Zekavat, S. M., & Bhatia, S. N. (2015). Mathematical framework for activity-based cancer biomarkers. Proceedings of the National Academy of Sciences of the USA , 112, 12627–12632.
Wang, Y.-C., & Chen, B.-S. (2011). A network-based biomarker approach for molecular investigation and diagnosis of lung cancer. BMC Medical Genomics, 4, 2.
Zeng, T., Sun, S. Y., Wang, Y., Zhu, H., & Chen, L. (2013). Network biomarkers reveal dysfunctional gene regulations during disease progression. FEBS Journal, 280, 5682–5695.
Vilar, S., Gonzalez-Diaz, H., Santana, L., & Uriarte, E. (2009). A network-QSAR model for prediction of genetic-component biomarkers in human colorectal cancer. Journal of Theoretical Biology, 261, 449–458.
von Loga, K., Woolston, A., Punta, M., Barber, L. J., Griffiths, B., Semiannikova, M., et al. (2020). Extreme intratumour heterogeneity and driver evolution in mismatch repair deficient gastro-oesophageal cancer. Nature Communications, 11, 139.
Ludwig, J. A., & Weinstein, J. N. (2005). Biomarkers in cancer staging, prognosis and treatment selection. Nature Reviews Cancer, 5, 845.
Chin, L., Andersen, J. N., & Futreal, P. A. (2011). Cancer genomics: From discovery science to personalized medicine. Nature medicine, 17, 297.
Simpson, R. J., Bernhard, O. K., Greening, D. W., & Moritz, R. L. (2008). Proteomics-driven cancer biomarker discovery: Looking to the future. Current Opinion in Chemical Biology, 12, 72–77.
Gal, S., Fidler, C., Lo, Y. M. D., Taylor, M., Han, C., Moore, J., et al. (2004). Quantitation of circulating DNA in the serum of breast cancer patients by real-time PCR. British Journal of Cancer, 90, 1211.
Sozzi, G., Conte, D., Mariani, L., Vullo, S. L., Roz, L., Lombardo, C., et al. (2001). Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Research, 61, 4675–4678.
Lehninger, A. L., Nelson, D. L., Cox, M. M., & Cox, M. M. (2005). Lehninger principles of biochemistry. New York: Macmillan.
Liu, P., Wang, Y., & Li, X. (2019). Targeting the untargetable KRAS in cancer therapy. Acta Pharmaceutica Sinica B. https://doi.org/10.1016/j.apsb.2019.03.002.
Pantsar, T. (2019). The current understanding of KRAS protein structure and dynamics. Computational and Structural Biotechnology Journal. https://doi.org/10.1016/j.csbj.2019.12.004.
Xi, X., Li, T., Huang, Y., Sun, J., Zhu, Y., Yang, Y., & Lu, Z. (2017). RNA biomarkers: Frontier of precision medicine for cancer. Non-coding RNA, 3, 9.
Berindan-Neagoe, I., Monroig, P. D. C., Pasculli, B., & Calin, G. A. (2014). MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA: A Cancer Journal for Clinicians, 64(311), 336.
Sand, M., Bechara, F. G., Gambichler, T., Sand, D., Bromba, M., Hahn, S. A., et al. (2016). Circular RNA expression in cutaneous squamous cell carcinoma. Journal of Dermatological Science, 83, 210–218.
Akao, Y., Nakagawa, Y., & Naoe, T. (2006). MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncology Reports, 16, 845–850.
Link, A., Balaguer, F., Shen, Y., Nagasaka, T., Lozano, J. J., Boland, C. R., & Goel, A. (2010). Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiology and Prevention Biomarkers, 19, 1766–1774.
Srinivas, P. R., Verma, M., Zhao, Y., & Srivastava, S. (2002). Proteomics for cancer biomarker discovery. Clinical chemistry, 48, 1160–1169.
Núñez, C. (2019). Blood-based protein biomarkers in breast cancer. Clinica Chimica Acta, 490, 113–127.
Ohno, Y., Maehashi, K., & Matsumoto, K. (2010). Label-free biosensors based on aptamer-modified graphene field-effect transistors. Journal of the American Chemical Society, 132, 18012–18013.
Li, L., Lei, Q., Zhang, S., Kong, L., & Qin, B. (2017). Screening and identification of key biomarkers in hepatocellular carcinoma: Evidence from bioinformatic analysis. Oncology Reports, 38, 2607–2618.
Homer, N., Szelinger, S., Redman, M., Duggan, D., Tembe, W., Muehling, J., et al. (2008). Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genetics, 4, e1000167.
Im, H. K., Gamazon, E. R., Nicolae, D. L., & Cox, N. J. (2012). On sharing quantitative trait GWAS results in an era of multiple-omics data and the limits of genomic privacy. American Journal of Human Genetics, 90, 591–598.
Schadt, E. E., Woo, S., & Hao, K. (2012). Bayesian method to predict individual SNP genotypes from gene expression data. Nature Genetics, 44, 603–608.
Hildebrand, P. W., Rose, A. S., & Tiemann, J. K. S. (2019). Bringing molecular dynamics simulation data into view. Trends in Biochemical Sciences. https://doi.org/10.1016/j.tibs.2019.06.004.
Karplus, M., & Petsko, G. A. (1990). Molecular dynamics simulations in biology. Nature, 347, 631.
Godwin, R. C., Melvin, R., & Salsbury, F. R. (2015). Molecular dynamics simulations and computer-aided drug discovery. In D. B. Singh (Ed.), computer-aided drug discovery (pp. 1–30). New York: Springer.
Klepeis, J. L., Lindorff-Larsen, K., Dror, R. O., & Shaw, D. E. (2009). Long-timescale molecular dynamics simulations of protein structure and function. Current Opinion in Structural Biology, 19, 120–127.
Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., et al. (2005). Scalable molecular dynamics with NAMD. Journal of Computational Chemistry, 26, 1781–1802.
Brooks, B. R., Brooks Iii, C. L., Mackerell, A. D., Jr., Nilsson, L., Petrella, R. J., Roux, B., et al. (2009). CHARMM: The biomolecular simulation program. Journal of Computational Chemistry, 30, 1545–1614.
Case, D. A., Cheatham Iii, T. E., Darden, T., Gohlke, H., Luo, R., Merz, K. M., Jr., et al. (2005). The Amber biomolecular simulation programs. Journal of Computational Chemistry, 26, 1668–1688.
Hess, B., Kutzner, C., Van Der Spoel, D., & Lindahl, E. (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4, 435–447.
Rampogu, S., Baek, A., Zeb, A., & Lee, K. W. (2018). Exploration for novel inhibitors showing back-to-front approach against VEGFR-2 kinase domain (4AG8) employing molecular docking mechanism and molecular dynamics simulations. BMC Cancer, 18, 264.
Jeevanandam, J., Tan, K. X., Danquah, M. K., Guo, H., & Turgeson, A. (2019). Advancing aptamers as molecular probes for cancer theranostic applications: The role of molecular dynamics simulation. Biotechnology Journal. https://doi.org/10.1002/biot.201900368.
Danquah, M. K., Guo, H.-B., Tan, K. X., & Bhakta, M. (2020). Atomistic probing of aptameric binding of CD19 outer membrane domain reveals an “aptamer walking” mechanism. Biotechnology Progress, 36, e2957.
Kumar, A., & Purohit, R. (2014). Use of long term molecular dynamics simulation in predicting cancer associated SNPs. PLoS Computational Biology, 10, e1003318.
Weako, J., Uba, A. I., Keskin, Ö., Gürsoy, A., & Yelekçi, K. (2020). Identification of potential inhibitors of human methionine aminopeptidase (type II) for cancer therapy: Structure-based virtual screening, ADMET prediction and molecular dynamics studies. Computational Biology and Chemistry, 86, 107244.
Reddy, P. S., & K. B. L., Shuchi Nagar, Vaddi Damodara Reddy, P. Sushma Murthy, K. Venkateswara Swamy, . (2018). Molecular Modeling, Docking, Dynamics and Simulation of Gefitinib and its Derivatives with EGFR in Non-small Cell Lung Cancer. Current Computer-Aided Drug Design, 14, 246–252.
Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31, 455–461.
Rich, R. L., & Myszka, D. G. (2004). Why you should be using more SPR biosensor technology. Drug Discovery Today: Technologies, 1, 301–308.
Suenaga, A., Ichikawa, M., Hatakeyama, M., Yu, X., Futatsugi, N., Narumi, T., et al. (2003). Molecular dynamics, free energy, and SPR analyses of the interactions between the SH2 domain of Grb2 and ErbB phosphotyrosyl peptides. Biochemistry, 42, 5195–5200.
Case, D. A., Darden, T. A., Cheatham, T. E., Simmerling, C. L., Wang, J., Duke, R. E., et al. (2019). Amber 11. San Francisco: University of California.
Amiri, M., Ajloo, D., Fazli, M., Mokhtarieh, A., Grivani, G., & Saboury, A. A. (2018). Spectroscopic, electrochemical, docking and molecular dynamics studies on the interaction of three oxovanadium (IV) Schiff base complexes with bovine serum albumin and their cytotoxicity against cancer. Journal of Biomolecular Structure and Dynamics, 36, 3753–3772.
Rao, N. V., Yoon, H. Y., Han, H. S., Ko, H., Son, S., Lee, M., et al. (2016). Recent developments in hyaluronic acid-based nanomedicine for targeted cancer treatment. Expert Opinion on Drug Delivery, 13, 239–252.
Bonifert, G., Folkes, L., Gmeiner, C., Dachs, G., & Spadiut, O. (2016). Recombinant horseradish peroxidase variants for targeted cancer treatment. Cancer Medicine, 5, 1194–1203.
Dong, K., Zhang, Y., Zhang, L., Wang, Z., Ren, J., & Qu, X. (2019). Facile preparation of metal−organic frameworks-based hydrophobic anticancer drug delivery nanoplatform for targeted and enhanced cancer treatment. Talanta, 194, 703–708.
Gavande, N. S., VanderVere-Carozza, P. S., Hinshaw, H. D., Jalal, S. I., Sears, C. R., Pawelczak, K. S., & Turchi, J. J. (2016). DNA repair targeted therapy: The past or future of cancer treatment? Pharmacology and Therapeutics, 160, 65–83.
Wang, H., & Mooney, D. J. (2018). Biomaterial-assisted targeted modulation of immune cells in cancer treatment. Nature Materials, 17, 761–772.
Jeevanandam, J., Pal, K., & Danquah, M. K. (2018). Virus-like nanoparticles as a novel delivery tool in gene therapy. Biochimie, 157, 38–47.
Jeevanandam, J., Sundaramurthy, A., Sharma, V., Murugan, C., Pal, K., Kodous, M. H. A., & Danquah, M. K. (2020). Sustainability of One-Dimensional Nanostructures: Fabrication and Industrial Applications. In G. Székely & A. G. Livingston (Eds.), Sustainable nanoscale engineering (pp. 83–113). New York: Elsevier.
Kuryk, L., Vassilev, L., Ranki, T., Hemminki, A., Karioja-Kallio, A., Levälampi, O., et al. (2017). Toxicological and bio-distribution profile of a GM-CSF-expressing, double-targeted, chimeric oncolytic adenovirus ONCOS-102: Support for clinical studies on advanced cancer treatment. PLoS ONE, 12, e0182715.
Massacesi, C., Di Tomaso, E., Urban, P., Germa, C., Quadt, C., Trandafir, L., et al. (2016). PI3K inhibitors as new cancer therapeutics: Implications for clinical trial design. OncoTargets and Therapy, 9, 203–210.
Mirzaei, H., Sahebkar, A., Salehi, R., Nahand, J., Karimi, E., Jaafari, M., & Mirzaei, H. (2016). Boron neutron capture therapy: Moving toward targeted cancer therapy. Journal of Cancer Research and Therapeutics, 12, 520–525.
Mishra, P., Nayak, B., & Dey, R. K. (2016). PEGylation in anti-cancer therapy: An overview. Asian Journal of Pharmaceutical Sciences, 11, 337–348.
Li, Y., Atkinson, K., & Zhang, T. (2017). Combination of chemotherapy and cancer stem cell targeting agents: Preclinical and clinical studies. Cancer Letters, 396, 103–109.
Belfiore, L., Saunders, D. N., Ranson, M., Thurecht, K. J., Storm, G., & Vine, K. L. (2018). Towards clinical translation of ligand-functionalized liposomes in targeted cancer therapy: Challenges and opportunities. Journal of Controlled Release, 277, 1–13.
Yeku, O., Zamarin, D., Gallagher, J., Aghajanian, C. A., & Konner, J. (2018). A phase II trial of TPIV200 (a polypeptide vaccine against folate receptor alpha) plus durvalumab (anti-PD-L1 antibody) in patients with platinum-resistant ovarian cancer. Gynecologic Oncology, 149, 56–57.
Fuller, S., Tomai, M., Sesay, M., Cunningham, D. & Price, J. (2017). Optimizing a unique cancer vaccine for intradermal delivery. BioPharm International Development Strategies for Emerging Therapies eBook.
Hill, A., Gotham, D., Fortunak, J., Meldrum, J., Erbacher, I., Martin, M., et al. (2016). Target prices for mass production of tyrosine kinase inhibitors for global cancer treatment. British Medical Journal Open, 6, e009586.
Meegan, M. J., & O’Boyle, N. M. (2019). Special issue “anticancer drugs.” Basel: Multidisciplinary Digital Publishing Institute.
Peiris, D., Spector, A. F., Lomax-Browne, H., Azimi, T., Ramesh, B., Loizidou, M., et al. (2017). Cellular glycosylation affects herceptin binding and sensitivity of breast cancer cells to doxorubicin and growth factors. Scientific Reports, 7, 43006.
Corrie, P. G., Terheyden, P., Ten Tije, A. J., Herbst, R., Jansen, R., Marples, M., et al. (2019). A prospective observational safety study of patients with BRAFV 600-mutated unresectable or metastatic melanoma treated with vemurafenib (Zelboraf Safety Study). British Journal of Dermatology, 180, 1254–1255.
Somlyai, G., Collins, T. Q., Meuillet, E. J., Hitendra, P., D’Agostino, D. P., & Boros, L. G. (2017). Structural homologies between phenformin, lipitor and gleevec aim the same metabolic oncotarget in leukemia and melanoma. Oncotarget, 8, 50187.
Parvizpour, S., Razmara, J., & Omidi, Y. (2018). Breast cancer vaccination comes to age: Impacts of bioinformatics. BioImpacts: BI, 8, 223.
Barbolosi, D., Ciccolini, J., Lacarelle, B., Barlési, F., & André, N. (2016). Computational oncology: Mathematical modelling of drug regimens for precision medicine. Nature Reviews Clinical Oncology, 13, 242.
Nalley, C. (2019). Utilizing computational oncology to better understand AML and MDS patients. Oncology Times, 41, 13.
Ding, M. Q., Chen, L., Cooper, G. F., Young, J. D., & Lu, X. (2018). Precision oncology beyond targeted therapy: Combining omics data with machine learning matches the majority of cancer cells to effective therapeutics. Molecular Cancer Research, 16, 269–278.
Bibault, J.-E., Giraud, P., & Burgun, A. (2016). Big data and machine learning in radiation oncology: State of the art and future prospects. Cancer Letters, 382, 110–117.
Majumder, J., Fine, J. A., Lantz, T. C., Conder, C. J., & Chopra, G. (2018). Abstract LB-076: Cancer cell specific lethality by degrading specific protein target network identified using a chemical screening based machine learning method. Cancer Research, 78, 076.
Wu, D., Yu, Y., Jin, D., Xiao, M.-M., Zhang, Z.-Y., & Zhang, G.-J. (2020). Dual-aptamer modified graphene field-effect transistor nanosensor for label-free and specific detection of hepatocellular carcinoma-derived microvesicles. Analytical Chemistry, 92, 4006–4015.
Mehmood, S., Khan, A. Z., Bilal, M., Sohail, A., & Iqbal, H. M. N. (2019). Aptamer-based biosensors: A novel toolkit for early diagnosis of cancer. Materials Today Chemistry, 12, 353–360.
Dehghani, S., Nosrati, R., Yousefi, M., Nezami, A., Soltani, F., Taghdisi, S. M., et al. (2018). Aptamer-based biosensors and nanosensors for the detection of vascular endothelial growth factor (VEGF): A review. Biosensors and Bioelectronics, 110, 23–37.
Hao, Z., Pan, Y., Huang, C., Wang, Z., & Zhao, X. (2019). Sensitive detection of lung cancer biomarkers using an aptameric graphene-based nanosensor with enhanced stability. Biomedical Microdevices, 21, 65.
Liu, J., Wei, T., Zhao, J., Huang, Y., Deng, H., Kumar, A., et al. (2016). Multifunctional aptamer-based nanoparticles for targeted drug delivery to circumvent cancer resistance. Biomaterials, 91, 44–56.
Ouyang, C., Zhang, S., Xue, C., Yu, X., Xu, H., Wang, Z., et al. (2020). Precision guided missile-like DNA nanostructure containing warhead and guidance control for aptamer-based targeted drug delivery into cancer cells in vitro and in vivo. Journal of the American Chemical Society. https://doi.org/10.1021/jacs.9b09782.
da Silva Luz, G. V., Barros, K. V. G., de Araújo, F. V. C., da Silva, G. B., da Silva, P. A. F., Condori, R. C. I., & Mattos, L. (2016). Nanorobotics in drug delivery systems for treatment of cancer: A review. Journal of Material Sciences and Engineering A, 6, 167–180.
Li, S., Jiang, Q., Liu, S., Zhang, Y., Tian, Y., Song, C., et al. (2018). A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nature Biotechnology, 36(3), 258
Hoop, M., Ribeiro, A. S., Rösch, D., Weinand, P., Mendes, N., Mushtaq, F., et al. (2018). Mobile magnetic nanocatalysts for bioorthogonal targeted cancer therapy. Advanced Functional Materials, 28, 1705920.
Ma, W., Zhan, Y., Zhang, Y., Shao, X., Xie, X., Mao, C., et al. (2019). An intelligent DNA nanorobot with in vitro enhanced protein lysosomal degradation of HER2. Nano Letters. https://doi.org/10.1021/acs.nanolett.9b01320.
Serrà, A., Vázquez-Mariño, G., García-Torres, J., Bosch, M., & Vallés, E. (2018). Magnetic actuation of multifunctional nanorobotic platforms to induce cancer cell death. Advanced Biosystem, 2, 1700220.
Jeevanandam, J., Aing, Y. S., Chan, Y. S., Pan, S., & Danquah, M. K. (2017). Nanoformulation and application of phytochemicals as antimicrobial agents. In A. Mehai (Ed.), Antimicrobial nanoarchitectonics: From synthesis to applications (Vol. 1, pp. 62–82). New York: Elsevier.
Jeevanandam, J., San Chan, Y., & Danquah, M. K. (2016). Nano-formulations of drugs: Recent developments, impact and challenges. Biochimie, 128, 99–112.
Jeevanandam, J., Chan, Y. S., Pan, S., & Danquah, M. K. (2019). Metal oxide nanocomposites: Cytotoxicity and targeted drug delivery applications. In K. Pal (Ed.), Hybrid nanocomposites: Fundamentals, synthesis and applications (pp. 111–147). Singapore: Pan Stanford Publishing.
Acknowledgements
The author (Dr. Jaison Jeevanandam) acknowledge the support of FCT-Fundação para a Ciência e a Tecnologia (Base Fund UIDB/00674/2020 and Programmatic Fund UIDP/00674/2020, Portuguese Government Funds), ARDITI-Agência Regional para o Desenvolvimento da Investigação Tecnologia e Inovação through the project M1420-01-0145-FEDER-000005-CQM+ (Madeira 14-20 Program).
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
JJ, GS, and KXT prepared the manuscript. JJ finalized the manuscript format and structure. MKD reviewed and revised the final manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jeevanandam, J., Sabbih, G., Tan, K.X. et al. Oncological Ligand-Target Binding Systems and Developmental Approaches for Cancer Theranostics. Mol Biotechnol 63, 167–183 (2021). https://doi.org/10.1007/s12033-020-00296-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-020-00296-2