Coincubation as miR-Loading Strategy to Improve the Anti-Tumor Effect of Stem Cell-Derived EVs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Renal Cancer Stem Cells Isolation and Culture
2.2. Human Liver Stem Cells Isolation and Culture
2.3. HLSC-EVs Isolation and Characterization
2.4. Electroporation Protocol
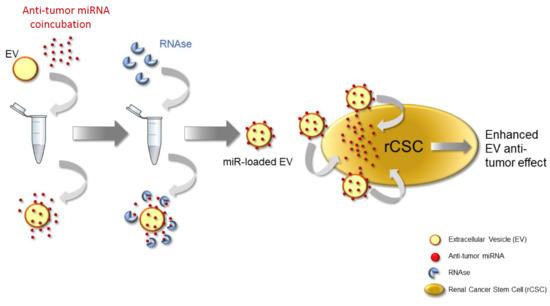
2.5. Coincubation Protocol
2.6. Cytofluorimetric EV Analysis
2.7. Apoptosis
2.8. Invasion
2.9. Super-Resolution Microscopy
2.10. EV Incorporation in Target Cells
2.11. miRNA Isolation and Real Time PCR
2.12. Protein Extraction and Western Blot
2.13. Statistical Analysis
3. Results
3.1. Comparison between Electroporation and Coincubation to Increase the EV Antitumor Effect
3.2. RNAse Treatment of Coincubated EVs
3.3. Anti-Tumor Effect and miR145 Transfer of RNase Treated Coincubated EVs
3.4. Protection of Surface Loaded miRNAs by RNA Binding Proteins
3.5. Coincubation Protocol Using Antitumor miRNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raposo, G.; Stahl, P.D. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019, 20, 509–510. [Google Scholar] [CrossRef] [PubMed]
- Wiklander, O.P.B.; Brennan, M.Á.; Lötvall, J.; Breakefield, X.O.; El Andaloussi, S. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019, 11, eaav8521. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.T.T.; Kavishka, J.M.; Zhang, D.X.; Pirisinu, M.; Le, M.T.N. Extracellular Vesicles as an Efficient and Versatile System for Drug Delivery. Cells 2020, 9, 2191. [Google Scholar]
- Massaro, C.; Sgueglia, G.; Frattolillo, V.; Baglio, S.R.; Altucci, L.; Dell’Aversana, C. Extracellular Vesicle-Based Nucleic Acid Delivery: Current Advances and Future Perspectives in Cancer Therapeutic Strategies. Pharmaceutics 2020, 12, 980. [Google Scholar] [CrossRef] [PubMed]
- Stremersch, S.; Vandenbroucke, R.E.; Van Wonterghem, E.; Hendrix, A.; De Smedt, S.C.; Raemdonck, K. Comparing exosome-like vesicles with liposomes for the functional cellular delivery of small RNAs. J. Control. Release 2016, 232, 51–61. [Google Scholar]
- Johnsen, K.B.; Gudbergsson, J.M.; Duroux, M.; Moos, T.; Andresen, T.L.; Simonsen, J.B. On the use of liposome controls in studies investigating the clinical potential of extracellular vesicle-based drug delivery systems—A commentary. J. Control. Release 2018, 269, 10–14. [Google Scholar]
- O’Loughlin, A.J.; Mäger, I.; de Jong, O.G.; Varela, M.A.; Schiffelers, R.M.; El Andaloussi, S.; Wood, M.J.A.; Vader, P. Functional Delivery of Lipid-Conjugated siRNA by Extracellular Vesicles. Mol. Ther. 2017, 25, 1580–1587. [Google Scholar]
- Devhare, P.B.; Ray, R.B. A novel role of exosomes in the vaccination approach. Ann. Transl. Med. 2017, 5, 23. [Google Scholar] [CrossRef]
- Sutaria, D.S.; Badawi, M.; Phelps, M.A.; Schmittgen, T.D. Achieving the Promise of Therapeutic Extracellular Vesicles: The Devil is in Details of Therapeutic Loading. Pharm. Res. 2017, 34, 1053–1066. [Google Scholar] [CrossRef] [Green Version]
- Pomatto, M.A.C.; Bussolati, B.; D’Antico, S.; Ghiotto, S.; Tetta, C.; Brizzi, M.F.; Camussi, G. Improved Loading of Plasma-Derived Extracellular Vesicles to Encapsulate Antitumor miRNAs. Mol. Ther. Methods Clin. Dev. 2019, 13, 133–144. [Google Scholar]
- Fonsato, V.; Collino, F.; Herrera, M.B.; Cavallari, C.; Deregibus, M.C.; Cisterna, B.; Bruno, S.; Romagnoli, R.; Salizzoni, M.; Tetta, C.; et al. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells 2012, 30, 1985–1998. [Google Scholar] [PubMed] [Green Version]
- Fonsato, V.; De Lena, M.; Tritta, S.; Brossa, A.; Calvetti, R.; Tetta, C.; Camussi, G.; Bussolati, B. Human liver stem cell-derived extracellular vesicles enhance cancer stem cell sensitivity to tyrosine kinase inhibitors through Akt/mTOR/PTEN combined modulation. Oncotarget 2018, 9, 36151–36165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopatina, T.; Grange, C.; Fonsato, V.; Tapparo, M.; Brossa, A.; Fallo, S.; Pitino, A.; Herrera-Sanchez, M.B.; Kholia, S.; Camussi, G.; et al. Extracellular vesicles from human liver stem cells inhibit tumor angiogenesis. Int. J. Cancer 2019, 144, 322–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brossa, A.; Fonsato, V.; Grange, C.; Tritta, S.; Tapparo, M.; Calvetti, R.; Cedrino, M.; Fallo, S.; Gontero, P.; Camussi, G.; et al. Extracellular vesicles from human liver stem cells inhibit renal cancer stem cell-derived tumor growth in vitro and in vivo. Int. J. Cancer 2020, 147, 1694–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bussolati, B.; Bruno, S.; Grange, C.; Ferrando, U.; Camussi, G. Identification of a tumor-initiating stem cell population in human renal carcinomas. FASEB J. 2008, 22, 3696–3705. [Google Scholar]
- Herrera, M.B.; Bruno, S.; Buttiglieri, S.; Tetta, C.; Gatti, S.; Deregibus, M.C.; Bussolati, B.; Camussi, G. Isolation and characterization of a stem cell population from adult human liver. Stem Cells 2006, 24, 2840–2850. [Google Scholar]
- Kooijmans, S.A.A.; Stremersch, S.; Braeckmans, K.; de Smedt, S.C.; Hendrix, A.; Wood, M.J.A.; Schiffelers, R.M.; Raemdonck, K.; Vader, P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Release 2013, 172, 229–238. [Google Scholar] [CrossRef]
- Hagiwara, K.; Katsuda, T.; Gailhouste, L.; Kosaka, N.; Ochiya, T. Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 2015, 589 Pt B, 4071–4078. [Google Scholar]
- Stewart, S.; Gessler, F.; Pluchino, S.; Moreau, K. Inside-out: Unpredicted Annexin A2 localisation on the surface of extracellular vesicles. Matters 2016, 10, 19185. [Google Scholar] [CrossRef]
- Mateescu, B.; Kowal, E.J.; van Balkom, B.W.; Bartel, S.; Bhattacharyya, S.N.; Buzás, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef] [Green Version]
- Fischer, S.; Cornils, K.; Speiseder, T.; Badbaran, A.; Reimer, R.; Indenbirken, D.; Grundhoff, A.; Brunswig-Spickenheier, B.; Alawi, M.; Lange, C. Indication of horizontal DNA gene transfer by extracellular vesicles. PLoS ONE 2016, 29, e0163665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelke, G.V.; Jang, S.C.; Yin, Y.; Lässer, C.; Lötvall, J. Human mast cells release extracellular vesicle-associated DNA. Matters 2016, 2, e201602000034. [Google Scholar] [CrossRef]
- Wahlgren, J.; Karlson, T.D.L.; Brisslert, M.; Vaziri Sani, F.; Telemo, E.; Sunnerhagen, P.; Valadi, H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012, 40, e130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slaby, O.; Redova, M.; Poprach, A.; Nekvindova, J.; Iliev, R.; Radova, L.; Lakomy, R.; Svoboda, M.; Vyzula, R. Identification of MicroRNAs associated with early relapse after nephrectomy in renal cell carcinoma patients. Genes Chromosomes Cancer 2012, 51, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Pirisinu, M.; Pham, T.C.; Zhang, D.X.; Hong, T.N.; Nguyen, L.T.; Le, M.T. Extracellular vesicles as natural therapeutic agents and innate drug delivery systems for cancer treatment: Recent advances, current obstacles, and challenges for clinical translation. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2020; Volume S1044-579X, p. 30179. [Google Scholar]
- O’Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.; O’Driscoll, L. miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S.; Takanashi, M.; Sudo, K.; Ueda, S.; Ishikawa, A.; Matsuyama, N.; Fujita, K.; Mizutani, T.; Ohgi, T.; Ochiya, T.; et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013, 21, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, H.; Bai, M.; Ning, T.; Ge, S.; Deng, T.; Liu, R.; Zhang, L.; Ying, G.; Ba, Y. Exosomes Serve as Nanoparticles to Deliver Anti-miR-214 to Reverse Chemoresistance to Cisplatin in Gastric Cancer. Mol. Ther. 2018, 26, 774–783. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Zhang, H.; Ning, T.; Liu, D.; Deng, T.; Liu, R.; Bai, M.; Zhu, K.; Li, J.; Fan, Q.; et al. Exosome-Delivered c-Met siRNA Could Reverse Chemoresistance to Cisplatin in Gastric Cancer. Int. J. Nanomed. 2020, 15, 2323–2335. [Google Scholar]
- Liang, G.; Kan, S.; Zhu, Y.; Feng, S.; Feng, W.; Gao, S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomed. 2018, 13, 585–599. [Google Scholar] [CrossRef] [Green Version]
- Kamerkar, S.; LeBleu, V.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar]
- Lou, G.; Song, X.; Yang, F.; Wu, S.; Wang, J.; Chen, Z.; Liu, Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharif, S.; Ghahremani, M.H.; Soleimani, M. Delivery of Exogenous miR-124 to Glioblastoma Multiform Cells by Wharton’s Jelly Mesenchymal Stem Cells Decreases Cell Proliferation and Migration, and Confers Chemosensitivity. Stem Cell Rev. Rep. 2018, 14, 236–246. [Google Scholar] [CrossRef]
- Tapparo, M.; Bruno, S.; Collino, F.; Togliatto, G.; Deregibus, M.C.; Provero, P.; Wen, S.; Quesenberry, P.J.; Camussi, G. Renal Regenerative Potential of Extracellular Vesicles Derived from miRNA-Engineered Mesenchymal Stromal Cells. Int. J. Mol. Sci. 2019, 20, 2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brossa, A.; Tapparo, M.; Fonsato, V.; Papadimitriou, E.; Delena, M.; Camussi, G.; Bussolati, B. Coincubation as miR-Loading Strategy to Improve the Anti-Tumor Effect of Stem Cell-Derived EVs. Pharmaceutics 2021, 13, 76. https://doi.org/10.3390/pharmaceutics13010076
Brossa A, Tapparo M, Fonsato V, Papadimitriou E, Delena M, Camussi G, Bussolati B. Coincubation as miR-Loading Strategy to Improve the Anti-Tumor Effect of Stem Cell-Derived EVs. Pharmaceutics. 2021; 13(1):76. https://doi.org/10.3390/pharmaceutics13010076
Chicago/Turabian StyleBrossa, Alessia, Marta Tapparo, Valentina Fonsato, Elli Papadimitriou, Michela Delena, Giovanni Camussi, and Benedetta Bussolati. 2021. "Coincubation as miR-Loading Strategy to Improve the Anti-Tumor Effect of Stem Cell-Derived EVs" Pharmaceutics 13, no. 1: 76. https://doi.org/10.3390/pharmaceutics13010076