Abstract
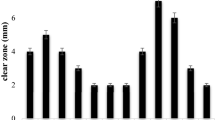
In the present study, twenty seven cellulose-degrading bacteria (CDB) were isolated from various organic manures and their cellulolytic activities were determined. The bacterial isolate CDB-26 showed the highest cellulolytic index, released 0.507 ± 0.025 mg/ml glucose and produced 0.196 ± 0.014 IU/ml cellulase enzyme under in vitro conditions. Biochemically, all the 27 isolates showed difference in the 6 biochemical tests performed. Further, all the 27 CDB isolates were subjected to various plant growth-promoting activities, and all CDB strains were positive for IAA production, GA3 production and siderophore production, whereas 19 strains were positive for ACC deaminase activity, 21 strains showed NH3 production and 19 strains were positive for HCN production. Out of 27 CDB isolates, 18 isolates were able to solubilize phosphate, 21 isolates were able to solubilize potash and 10 CDB isolates were found positive for silica solubilization. The molecular diversity among different CDB isolates was studied through ARDRA and demonstrated very high genetic diversity among these bacteria. The in vitro cellulose-degradation potential of these CDB isolates using vegetable waste as substrate were also assessed, and the 3 CDB isolates viz. Serratia surfactantfaciens (CDB-26), Stenotrophomonas rhizophila (CDB-16) and Pseudomonas fragi (CDB-5) showed the highest cellulose-degrading potential under in vitro conditions. Hence, the cellulolytic microbes isolated in the present study could be used for effective bioconversion of plant biomasses into enriched compost.



Similar content being viewed by others
Abbreviations
- PCR–RFLP:
-
Polymerase chain reaction–restriction fragment length polymorphism
- ARDRA:
-
Amplified 16S ribosomal DNA restriction analysis
- UPGMA:
-
Unweighted pair group method with arithmetic mean
References
Shankar TJ, Sokhansanj S, Hess JR, Wright CT, Boardman RD (2011) A review on biomass torrefaction process and product properties for energy applications. Ind Biotechnol 7(5):384–401
Pan I, Dam B, Sen SK (2012) Composting of common organic wastes using microbial inoculants. 3 Biotech 2(2):127–134
Balachandrababu A, Revathi MM, Yadav A, Sakthivel N (2012) Purification and characterization of a thermophilic cellulose from a novel cellulolytic strain, Paenibacillus barcinonensis. J Microbiol Biotechnol 22:1501–1509
Sreeja SJ, Jeba MP, Sharmila JF, Steffi T, Immanuel G, Palavesam A (2013) Optimization of cellulase production by Bacillus altitudinis APS MSU and Bacillus licheniformis APS2 MSU, gut isolates of fish Etroplus suratensis. Int J Adv Res Technol 2(4):401–406
Lederberg J (1992) Cellulases: encyclopaedia of microbiology. Academic Press, Cambridge, pp 1–5
Lynd LR, Weimer PJ, Van WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
Wright SF (2003) The important of soil microorganisms in aggregate stability. North Central Ext Ind Soil Fertility Conf 19:93–98
Aubert JP, Beguin P, Millet J (1987) Biochemistry and genetics of cellulose degradation, fungal and bacterial enzyme systems and their manipulation. FEMS Symp 43:11–30
Immanuel G, Dhanusha R, Prema P, Palavesam A (2006) Effect of different growth parameters on endoglucanase enzyme activity by bacteria isolated from coir retting effluents of estuarine environment. Int J Environ Sci Technol 3:25–34
Muhammad I, Safdar A, Quratulain S, Muhammad N (2012) Isolation and screening of cellulolytic bacteria from soil and optimization of cellulase production and activity. J Biochem 37(3):287–293
Mahjabeen F (2016) Isolation of cellulolytic bacteria from soil and identification by 16s rRNA gene sequencing. Bangladesh J Microbiol 33:23–44
Miller GL (1959) Use of dinitrosalicylic acid reagent for the determination of reducing sugar. Anal Chem 31(3):426–428
Kour R, Jain D, Bhojiya AA, Sukhwal A, Sanadhya S, Saheewala H, Jat G, Singh A, Mohanty SR (2019) Zinc biosorption, biochemical and molecular characterization of plant growth-promoting zinc-tolerant bacteria. 3 Biotech 9:421
Jain D, Sanadhya S, Saheewala H, Maheshwari D, Shukwal A, Singh PB, Meena RH, Choudhary R, Mohanti SR, Singh A (2020) Molecular diversity analysis of plant growth promoting rhizobium isolated from groundnut and evaluation of their field efficacy. Curr Microbiol 77:1550–1557
Jain D, Sunda SD, Sanadhya S, Dhruba JN, Khandelwal SK (2017) Molecular characterization and PCR-based screening of cry genes from Bacillus thuringiensis strains. 3 Biotech 7:4
Cihan CA, Tekin N, Ozcan B, Cokmus C (2012) The genetic diversity of genus Bacillus and the related genera revealed by 16s rRNA gene sequences and ARDRA analyses isolated from geothermal regions of turkey. Braz J Microbiol 43(1):309–324
Rohlf FJ (1998) NTSYSpc numerical taxonomy and multivariate analysis system version 2.0 user guide. Applied Biostatistics Inc., Setauket, NY, p 37
Gautam SP, Bundela PS, Pandey AK, Jamaluddin AMK, Sarsaiya S (2012) Diversity of cellulolytic microbes and the biodegradation of municipal solid waste by a potential strain. Int J Microbiol 2012:1–12
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Thompson JD, Higgins DG, Gibson J (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res 22(22):4673–4680
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Ahmad M, Zahir ZA, Khalid M (2013) Efficacy of Rhizobium and Pseudomonas strains to improve physiology, ionic balance and quality of mung bean under salt-affected conditions on farmer’s fields. Plant Physiol Biochem 63:170–176
Eida MF, Nagaoka T, Wasaki J, Kouno K (2012) Isolation and characterization of cellulose-decomposing bacteria inhabiting sawdust and coffee residue composts. Microbes Environ 27(3):226–233
De Marco ÉG, Heck K, Martos ET, Van der Sand ST (2017) Purification and characterization of a thermostable alkaline cellulase produced by Bacillus licheniformis380 isolated from compost. Anais da Acad Brasil de Ciências 89(3 Suppl.):2359–2370
Rawway M, Ali SG, Badawy AS (2017) Isolation and identification of cellulose degrading bacteria from different sources at Assiut governorate (Upper Egypt). J Ecol Health Environ 6(1):15–24
Gupta P, Samant K, Sahu A (2012) Isolation of cellulose degrading bacteria and determination of their cellulolytic potential. Int J Appl Microbiol Sci 1(2):13–23
Ferbiyanto A, Rusmana I, Raffiudin R (2015) Characterization and identification of cellulolytic bacteria from gut of worker Macrotermes gilvus. HAYATI J Biosci 22:197–200
Ramalingam K, Ramasamy G (2013) Isolation, screening and characterization of cellulase producing Bacillus subtilis KG10 from virgin forest of KovaiKutralam, Coimbatore, India. Res J Biotechnol 8(6):17
Deka D, Bhargav P, Sharma A, Goyal D, Jawed M, Goyal A (2011) Enchanment of cellulase activity from a new strain of Bacillus subtilis by medium optimization and analysis with various cellulosic substrates. Enzyme Res 151656:1–8. https://doi.org/10.4061/2011/151656
Singh S, Moholkar VS, Goyal A (2013) Isolation identification and characterization of a cellulolytic Bacillus amyloliquefaciens strain SS35 from rhinoceros dung. Int Sch Res Net (ISRN) Microbiol 2013:1–7
Rastogi G, Muppidi GL, Gurram RN, Adhikari A, Bischoff KM, Hughes SR, Sani RK (2009) Isolation and characterization of cellulose-degrading bacteria from the deep subsurface of the Homestake gold mine, Lead, South Dakota, USA. J Ind Microbiol Biotechnol 36(4):585–598
Handique G, Phukan A, Bhattacharyya B, Baruah A, Rahman SW, Baruah R (2017) Characterization of cellulose degrading bacteria from the larval gut of the white grub beetle Lepidiota mansueta (Coleoptera: Scarabaeidae). Arch Insect Biochem Physiol 94:e21370
Islam F, Roy N (2018) Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC Res Notes 11:445
Weselowski B, Nathoo N, Eastman AW, MacDonald J, Yuan Z (2016) Isolation, identification and characterization of Paenibacillus polymyxa CR1 with potentials for biopesticide, biofertilization, biomass degradation and biofuel production. BMC Microbiol 16:244
Passari AK, Mishra VK, Leo VV, Gupta VK, Singh BP (2016) Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol Res 193:57–73
Szymańska S, Płociniczak T, Piotrowska-Seget Z, Złoch M, Ruppel S, Hrynkiewicz K (2016) Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiol Res 182:68–79
Wirth S, Ulrich A (2002) Cellulose-degrading potentials and phylogenetic classification of carboxymethyl-cellulose decomposing bacteria isolated from soil. Syst Appl Microbiol 25:584–591
Huang S, Sheng P, Zhang H (2012) Isolation and Identification of Cellulolytic Bacteria from the Gut of Holotrichia parallela Larvae (Coleoptera: Scarabaeidae). Int J Mol Sci 13:2563–2577
Sarkar P, Chourasia R (2017) Bioconversion of organic solid wastes into biofortified compost using a microbial consortium. Int J Recycl Org Waste Agric 6(4):321–334
Al-Dhabi NA, Esmail GA, Ghilan A-KM, Arasu MV (2019) Composting of vegetable waste using microbial consortium and biocontrol efficacy of Streptomyces Sp. Al-Dhabi 30 isolated from the Saudi Arabian environment for sustainable agriculture. Sustainability 11:6845
Dantur KI, Enrique R, Welin B, Castagnaro AP (2015) Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express 5:15
Sun S, Zhang Y, Liu K, Chen X, Jiang C, Huang M, Zang H, Li C (2020) Insight into biodegradation of cellulose by psychrotrophic bacterium Pseudomonas sp. LKR-1 from the cold region of China: optimization of cold-active cellulase production and the associated degradation pathways. Cellulose 27:315–333
Sethi S, Datta A, Gupta BL, Gupta S (2013) Optimization of cellulase production from bacteria isolated from soil. Int Sch Res Net (ISRN) Biotechnol. https://doi.org/10.5402/2013/985685
Acknowledgements
The financial assistance from All India Network Project on soil biodiversity and bio-fertilizers and Rastriya Krishi Vikas Yojana (RKVY) research project are highly acknowledged. All India Network Project on Organic farming is gratefully acknowledged for providing freshly prepared organic manures. The support of Dean, RCA and Director of Research, MPUAT is also acknowledged.
Author information
Authors and Affiliations
Contributions
DJ, SRM conceived and designed the experiments; R, AAB, SC performed laboratory experiments; DJ and DR wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jain, D., Ravina, Bhojiya, A.A. et al. Polyphasic Characterization of Plant Growth Promoting Cellulose Degrading Bacteria Isolated from Organic Manures. Curr Microbiol 78, 739–748 (2021). https://doi.org/10.1007/s00284-020-02342-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02342-3