Abstract
Many studies have established the functional properties of Lacticaseibacillus rhamnosus GG, previously known as Lactobacillus rhamnosus GG, marketed worldwide as a probiotic. The extraordinary capacity of L. rhamnosus GG to bind to human mucus and influence the immune system especially stand out. Earlier, we have shown the key role of its SpaCBA sortase-dependent pili encoded by the spaCBA-srtC1 gene cluster herein. These heterotrimeric pili consist of a shaft pilin SpaA, a basal pilin SpaB, and tip pilin SpaC that contains a mucus-binding domain. Here, we set out to characterize a food-grade non-GMO mutant of L. rhamnosus GG, strain PA11, which secretes its pilins, rather than coupling them to the cell surface, due to a defect in the housekeeping sortase A. The sortase-negative strain PA11 was extensively characterized using functional genomics and biochemical approaches and found to secrete the SpaCBA pili into the supernatant. Given the functional importance and uniqueness of the mucus-binding pili of L. rhamnosus GG, strain PA11 offers novel opportunities towards the characterization and further therapeutic application of SpaCBA pili and their low-cost, large-scale production.
Key points
•Creation of pilus-secreting mutant (PA11) of the key probiotic LGG.
•Strain PA11 is defective in a functional housekeeping sortase SrtA.
•Strain PA11 opens novel biotherapeutic application avenues.
Graphical abstract

Similar content being viewed by others
Introduction
Lacticaseibacillus rhamnosus GG (previously known as Lactobacillus rhamnosus GG) (Zheng et al. 2020) is a widely marketed lactic acid bacterium (commercialized as LGG®) that has been extensively studied for its probiotic properties for decades (Saxelin et al. 2005). Among others, the beneficial effect of L. rhamnosus GG has been demonstrated in the prevention of diarrhea in children and adults (Arvola et al. 1999; Wolvers et al. 2010; Szajewska and Kolodziej 2015) and respiratory tract infections in children (Hatakka et al. 2001; Kumpu et al. 2012; Luoto et al. 2014). This effect has been mainly associated to the high adhesion capacity of L. rhamnosus GG attributed to the presence of unique mucus-binding pili on its surface, which are absent in closely related strains, such as L. rhamnosus LC705 (Kankainen et al. 2009). These sortase-dependent pili are heterotrimeric proteinaceous cell wall appendages involved in adherence, biofilm formation, and host signaling (Kankainen et al. 2009; von Ossowski et al. 2010; Lebeer et al. 2012; Ardita et al. 2014; Chandrasekharan et al. 2019).
The biogenesis of the L. rhamnosus GG pili is encoded by the spaCBA-srtC1 gene cluster (Kankainen et al. 2009). The pilus shaft is built out of the major pilin SpaA, while the SpaB pilin terminates pilus synthesis (Reunanen et al. 2012; Douillard et al. 2014). The pilus shaft and tip are decorated with a mucin-binding domain containing SpaC subunit (Reunanen et al. 2012). The latter subunit is the major contributor to the strong mucus adhesion phenotype of L. rhamnosus GG (Kankainen et al. 2009) by specifically binding to the non-reducing termini of mucus glycoconjugates (Nishiyama et al. 2016). More in particular, the SpaC pilin harbors a C-terminal collagen-binding domain and an N-terminal von Willebrand factor type A domain (Tripathi et al. 2013). Especially, the latter one is thought to contribute to the extraordinary adhesion capacities of the SpaC pilin and by extension of the SpaCBA pili and L. rhamnosus GG cells (Tripathi et al. 2013; Kant et al. 2016). The different pilin subunits are polymerized to form the pilus, with lengths of over 1 μm by a pilin-specific transpeptidase sortase SrtC1, which recognizes highly conserved LPxTG and YPKN motifs and covalently attached to the cell wall by a housekeeping sortase, SrtA (Hendrickx et al. 2011; Reunanen et al. 2012). The action of both sortases during assembly and anchoring of the pilus is further determined by either a triple or single glycine motif recognized by the pilin-specific sortase and housekeeping sortase of L. rhamnosus GG, respectively (Douillard et al. 2014). Further investigation of the pilin-specific SrtC1 revealed an N-terminal GYPSY domain important for its functionality (Douillard et al. 2016). The intensive study of these peculiar pili of L. rhamnosus GG has already generated important insights in pilus assembly and function in Gram-positive species in general. Sortase-dependent pili are key cell wall appendages of Gram-positive species, especially in pathogens, where they are virulence factors (Mandlik et al. 2008). In earlier work, we uncovered a striking structural and functional resemblance between the pili of the probiotic L. rhamnosus GG and the nosocomial pathogen Vancomycin-resistant Enterococcus (VRE), showing that SpaCBA pili can drive competitive exclusion of VRE (Tytgat et al. 2016a).
To get insight into the genes governing SpaCBA pilus formation and to produce a panel of strains that can be safely used in therapeutic applications, we earlier set out to produce and characterize non-piliated derivatives of L. rhamnosus GG using a chemical mutagenesis approach (Rasinkangas et al. 2014). Since random chemical mutagenesis was used to produce these, the resulting L. rhamnosus GG derivatives are allowed to be used in human trials, as they are not considered to be genetically modified organisms (GMO) and can be used in food applications in the EU, that is known for its strict regulation (EU 2001). In this food-grade mutagenesis campaign, we also obtained strain PA11 from L. rhamnosus GG that contains a critical mutation in the house-keeping sortase (srtA) gene and was found to secrete the mucus-binding SpaC-harboring pili. In this work, we report on the in-depth characterization of L. rhamnosus PA11 at the phenotypic, biochemical, and genomic level.
Materials and methods
Bacterial strains and growth conditions
Lacticaseibacillus rhamnosus GG (ATCC 53103) and Lacticaseibacillus rhamnosus LC705 were obtained from Valio Ltd., Helsinki, Finland. The previously characterized L. rhamnosus GG PB12 strain (Rasinkangas et al. 2014) and closely related L. rhamnosus LC705 were included as SpaCBA pilus-less controls in experiments. All strains were grown in De Man-Rogosa-Sharpe (MRS) medium and agar (Difco) at 37 °C.
Mutagenesis
Mutagenesis was performed as described earlier in Rasinkangas et al. (Rasinkangas et al. 2014) using 2% (v/v) ethyl methanesulphonate (EMS; Sigma Aldrich). EMS is an alkylating mutagen and produces mainly transition mutations (Sega 1984; Parekh et al. 2000). The L. rhamnosus GG derivative strain PA11 was isolated based on a previously described enrichment procedure where we selected for mutants that were reduced in the binding to mucus in microtiter plates (Rasinkangas et al. 2014). Strain PA11 was obtained among a dozen of pilus-less mutants and stood out because it did not bind mucus, but still produced pili. L. rhamnosus PA11 is available under number DSM 111733 at the DSM-Z Braunschweig, Germany.
Immunoelectron microscopy
Immunoelectron microscopy double labeling was performed as described previously (Reunanen et al. 2012; Rasinkangas et al. 2014), with antisera against SpaA and SpaC pilins (Douillard et al. 2016; Tytgat et al. 2016b). SpaA was labeled with 10 nm, and SpaC was labeled with 5 nm protein A-gold particles. The samples were visualized using JEM-1400 transmission electron microscope (JEOL).
Quantification of surface-located pilins using immunofluorescence
Immunofluorescence labeling, quantification, and microscopy of pilins on the surface of cells were performed essentially as described previously in Rasinkangas et al. (2014). Initial labeling was performed using SpaA and SpaC antisera followed by secondary labeling with Alexa Fluor 488-labeled goat anti-rabbit antibody (Invitrogen). To normalize the fluorescence intensity for SpaA and SpaC to the total quantity of cells in the samples, chromosomal DNA of the cells was labeled with 1:1000-diluted 4′,6-diamidino-2-phenylindole (DAPI, Dilactate-form, Thermo Fisher Scientific) at the same time with the secondary antibody labeling. Fluorescence intensities of freshly labeled samples were measured with a Victor3 1460 multilabel counter (Perkin Elmer) using separate measurement programs for each dye. The fluorescence intensity results from the immunofluorescence labeling were divided by the corresponding DAPI intensity to obtain a normalized fluorescence value for each strain.
Mucus adherence measurements
Mucus adherence of the strains was measured as described previously (Vesterlund et al. 2005; Rasinkangas et al. 2014). In short, strains were grown overnight in the presence of 3H-labeled thymidine (Perkin Elmer). These cultures were then incubated in a Maxisorp microtiter plate coated with porcine type II mucus (Sigma). Wells were washed with PBS (Oxoid) and the remaining adherent cells were lysed. Radioactivity of the samples was measured with a Wallac 1480 Wizard 3 automatic gamma counter (Perkin Elmer). Three repeats of the mucus binding assay were performed, and in each assay, 3–6 technical replicates were prepared for each strain. Data analysis was performed using GraphPad Prism® 8. Significant differences between two groups were calculated using unpaired t tests, and the significance level was set at p < 0.0001.
Genome sequencing
Extraction of genomic DNA, shotgun genome sequencing (Illumina, MiSeq), and processing of sequences were performed as described earlier (Rasinkangas et al. 2014). Sequence assemblies were aligned to the L. rhamnosus GG genome (Kankainen et al. 2009) using MUMmer 3.0 software (Kurtz et al. 2004) as described previously. The mutation detected in the srtA gene of the PA11 strain was further verified with Sanger sequencing. Genomic DNA was also sent to BaseClear (Leiden, NL), where single-molecule genome sequencing (PacBio) was performed and analyzed according to their procedures (paired-end sequencing on Illumina HiSeq2500 system followed by variant detection).
Western blot analysis of the cell wall-located pilins
Cell wall proteins were extracted as detailed previously (Avall-Jaaskelainen et al. 2003; Rasinkangas et al. 2014) and analyzed with Western blot, as described in Rasinkangas et al. (Rasinkangas et al. 2014). The optical density of the cultures was adjusted to A600 = 8.0 before protein extraction, and the protein extracts were diluted 1:300 for detection with SpaA antiserum and 1:200 for SpaC Western blot. Secreted proteins were analyzed by blotting a 10.5-μl sample from the culture medium.
Secretome
The supernatant of a 500-ml culture of L. rhamnosus GG wild type and PA11 at OD600 7 and was collected and filtered over a 0.45-μm filter. After TCA precipitation (20% (w/v) final concentration, Sigma), the pellet was washed twice with ice-cold acetone. Dried samples were resuspended in 200 uL 8% (w/v) SDS and further in 400 uL ddH2O, using sonication in a water bath to dissolve the samples. Samples were run on an 8% SDS gel after which the whole lane was cut into ten pieces and prepared for further analysis by nanoUPLC-MS/MS. The gel pieces were processed as described earlier (Lee et al. 2003). In brief, the gels were reduced and alkylated by resp. 10 mM dithiothreitol and 20 mM iodoacetamide. The gel pieces were washed twice in 50% (v/v) acetonitrile in 50 mM ammonium bicarbonate buffer pH 8.5, followed by a wash with 100% (v/v) acetonitrile. Gel pieces were dried by SpeedVac and incubated with 0.1 μg of trypsin overnight. Tryptic peptides were desalted by C18 Zip-Tip (Millipore, USA) and dried by SpeedVac. Twenty percent of the dissolved peptide sample was analyzed using an Easy-nLC™ 1000 system (Thermo Scientific) coupled to a LTQ-Orbitrap Fusion mass spectrometer (Thermo Scientific). Peptides were resuspended in 2.5% (v/v) acetonitrile (ACN) with 0.1% (v/v) formic acid (FA) and loaded on a self-made fritted column (75 μm × 150 mm) packed with reverse phase C18 material AQ, 3 μm 200 Å, (Bischoff GmbH, Leonberg, Germany) and eluted by a gradient (from 5 to 35% of solution B (99.9% ACN, 0.1% FA) for 30 min, 55% of B for 10 min, 97% of B for 10 min, with a flow rate of 300 nl/min). One scan cycle included a full-scan MS survey spectrum, followed by sequential HCD MS/MS on the most intense signals (> 50,000) to the maximal cycle time for 3 s. Full-scan MS spectra (400–2000 m/z) and HCD MS/MS spectra were recorded in the FT-Orbitrap (resolution of 120,000 at 400 m/z for MS and 60,000 at 400 m/z for MS/MS). HCD was performed with a target value of 5e4 and stepped collision energy rolling from 35 NCE was applied. For full Fourier transform MS, AGC target values were 2e5. Dynamic exclusion with a single repeat count, 15 s repeat duration, and 60 s exclusion duration was used for all experiments. The data was transformed to mgf file format by Proteome Discoverer 2.0 (Thermo, USA) and searched against the L. rhamnosus GG proteome (Kankainen et al. 2009) using the Mascot search engine. Quantification was performed by Progenesis QI.
Results
Following up on our previous work selecting and characterizing non-mucus binding non-GMO derivatives of the model probiotic L. rhamnosus GG, we report on the isolation and characterization of the PA11 derivative that was found to secrete its SpaCBA pili.
Selection of pili-secreting L. rhamnosus GG derivative
Using ethyl methanesulfonate (EMS) as a chemical mutagenesis agent, derivatives of L. rhamnosus GG (ATCC 53103) were generated and non-anti-SpaC-binding mutants were enriched and further characterized (Rasinkangas et al. 2014). One isolate, strain PA11, in particular, stood out during the characterization of the non-mucus adherent strains as it appeared to secrete its pilins, which triggered a more in-depth investigation.
Immunoelectron microscopy shows that PA11 secretes its pili
We used immunoelectron microscopy (immuno-EM) to reveal if and how the pili of strain PA11 are displayed on the cells. Cells were treated both with 10 nm gold particles coupled with anti-SpaA (shaft pilin) antibodies and 5 nm gold particles coupled to anti-SpaC (tip pilin) antibodies. Labeling of the wild-type strain showed co-labeling of both particles on the proteinaceous appendages on the cell surface, while labeling was absent in the non-piliated PB12 strain we described earlier (Fig. 1a) (Rasinkangas et al. 2014). The cells of strain PA11 were practically non-piliated, only showing minor amounts of pili on the cell surface. In contrast to the picture of strain PB12, the bulk of the pili of strain PA11 appeared to be loosely secreted in the milieu surrounding the cells, as evident by the interaction of the gold particles with these secreted pili (Fig. 1a).
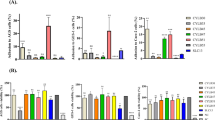
Phenotypical characterization of the pilus-secreting L. rhamnosus GG derivative PA11. a Immunoelectron micrographs of SpaC-SpaA-double labeling of L. rhamnosus strains. SpaC is labeled with 5 nm gold particles, while SpaA is labeled with 10 nm gold particles. A representative figure is shown for L. rhamnosus GG, the pilus-deficient PB12, and the pilus-secreting derivative PA11. b SpaA and SpaC immunofluorescence labeling. The strains were labeled with SpaA and SpaC antisera, and the antibodies were detected with Alexa 488-labeled secondary antibody. c SpaA and SpaC immunofluorescence labeling quantitative results. DAPI-normalized fluorescence intensity measurement results. Results are normalized to values for the parental strain both for SpaA and SpaC quantification. A representative experiment is shown. d Adherence of the different strains to porcine mucus. Wild-type L. rhamnosus GG was included as positive control, while the closely related, but SpaCBA-negative strain, L. rhamnosus LC705 and the non-piliated derivative PB12 were included as negative controls. The result is the average of 3 biological measurements, in which 3–6 technical repeats were included. Standard deviation is depicted
Quantification of pilins using immunofluorescence labeling
In order to confirm the immuno-EM observations and quantify the amount of pili produced by each strain, we performed immunofluorescence assays. Fluorescence of each sample was normalized to DAPI staining of nucleic acids, and all values were compared to the wild-type strain. In line with the immuno-EM observations, the quantity of SpaA and SpaC pilins at the surface of the pilus-secreting PA11 strain was lower compared to the parental strain (with a normalized fluorescence intensity for SpaA of 0.73, and for SpaC of 0.47), albeit slightly higher than in the pilus-deficient strain PB12 (SpaA 0.1, SpaC 0.2) (Fig. 1b, c). The apparent absence of SpaC staining of strain PA11 is in accordance with its selection process, as mutants were selected based on their non-interaction with anti-SpaC antibodies (Fig. 1b) (Rasinkangas et al. 2014).
Strain PA11 is deficient in mucus adhesion
Given the microscopical observation suggesting that strain PA11 secretes its pili, a mucus adhesion experiment was set up to investigate how this affected the ability of PA11 to interact with mucus. Mucus adhesion to porcine mucus was significantly reduced (p < 0.0001) in the pilus-secreting derivative PA11 (6.4% of the cells, SEM 0.6) compared to the parental strain (41.7%, SEM 1.7) (Fig. 1d). The pilus-less strain PB12 and the closely related L. rhamnosus LC705, which is deficient in the spaCBA-srtC1 genes (Kankainen et al. 2009), were included as negative controls (respectively 0.58%, SEM 0.18 and 0.77%, SEM 0.15). Our results further corroborate the importance of SpaCBA pili in the strong mucus adherence phenotype of L. rhamnosus GG (Kankainen et al. 2009; von Ossowski et al. 2010; Lebeer et al. 2012).
Genome analysis reveals a mutation in the housekeeping sortase SrtA
To investigate the genotype underlying our phenotypic observations on the secretion of pili into the growth medium by strain PA11, we performed deep shotgun (Illumina MiSeq, 142-fold coverage, 130 Mbp, Rasinkangas et al. 2014) as well as single-molecule (PacBio, BaseClear, Leiden, NL) sequencing of its genome (Supplementary File 1). The genome sequence of L. rhamnosus GG-derived strain PA11 can be found in the NCBI Sequence Read Archive (SRA) database with accession number SAMN04440354, and all sequencing data was deposited as part of BioProject PRJNA309744. In contrast to the earlier characterized non-piliated derivatives of L. rhamnosus GG (Rasinkangas et al. 2014), the spaCBA-srtC1 gene cluster responsible for SpaCBA pili production was not affected. A total of 86 SNPs were detected with shotgun sequencing and 103 SNPs in 87 genes with single-molecule sequencing, including a SNP in the srtA gene encoding the housekeeping sortase SrtA, LGG_RS10305 (LGG_02143) (Supplementary File 1). The mutation detected in the srtA gene of the PA11 strain was further verified with Sanger sequencing and included a point mutation changing the G at position 632 to an A, resulting in a change of amino acid residue at position 211 from Arg to Gln (Supplementary File 1; Fig. 2).
Multiple sequence alignment of SrtA sequences. The protein sequence of L. rhamnosus GG SrtA was aligned against members of the SrtA protein family, cd06165. The catalytic triad is indicated with a number sign, and the TXTLC catalytic motif is underlined. The mutation of residue 211 in the PA11 strain of an Arg into a Gln, resulted in the substitution of one of the three amino acids of the catalytic triad: His, Cys, and Arg
Western blot analysis points towards secretion of mature pili by PA11
Based on the genotypical and phenotypical results described above, we set out to further strengthen the observation that strain PA11 secretes most of its mature pilins into its environment. Cell wall-associated fractions from wild-type L. rhamnosus GG and its derivative strain PA11 were analyzed by Western blot to visualize potential differences in the pilus ladder, typically ranging in the high molecular weight regions (Kankainen et al. 2009) (Fig. 3). In accordance with the immuno-EM observations (Fig. 1a), only pili of lower molecular weight seemed to be present on the cell surface of PA11, while the higher molecular weight pili seemed to be secreted into the culture medium (Fig. 3). This further supports the hypothesis that pili of mature length produced by PA11 are released in the medium because of the lack of sortase A activity, due to the mutation in the catalytic triad of SrtA. Work in Staphylococcus aureus, which like L. rhamnosus is a Gram-positive bacterium, showed the same geno-phenotype relationship, with abolishment of the Arg residue resulting in very strong, but not full, reduction of SrtA functionality (Marraffini et al. 2004).
Western blot analyses of PA11 supernatant and cell wall protein fractions. Both SpaA and SpaC blots are shown for cell wall (CW) and supernatant (SN) protein fractions of L. rhamnosus GG and PA11. The cell wall extracts were extracted as detailed previously (Avall-Jaaskelainen et al. 2003; Rasinkangas et al. 2014) from cultures with an adjusted optical density of A600 = 8.0. Samples were diluted 1:300 for the SpaA and 1:200 for the SpaC blot. To analyze proteins secreted by both strains, a 10-μl aliquot of the cell culture medium was blotted
Analysis of the secreted proteome confirms increased secretion of pili by PA11
The proteome of secreted proteins of strain PA11 and wild-type L. rhamnosus GG was determined to investigate if the SrtA defect also affected other proteins beyond the SpaCBA pili. Secretome analysis resulted in 27 proteins that were enriched in the supernatant of the PA11 strain compared to the parental strain (Table 1; Supplementary File 2). Of interest, seven of these stood out as they made up the majority (over 75%) of the peptide counts. All of these secreted proteins harbored an LPXTG-motif and thus normally relied on SrtA activity for their anchoring to the cell surface (indicated in Table 1) (Douillard et al. 2014). The most abundant peptide counts derived from the large adhesin exoprotein of unknown function (LGG_RS13990) and the lactocepin protein (LGG_RS13070), a putative protease that in L. casei was shown to have potent immunomodulatory properties (von Schillde et al. 2012). In conclusion, the secretion of sortase-dependent proteins due to the defect in SrtA in the PA11 strain, further illustrates the importance of SrtA in anchoring LPXTG-harboring proteins on the cell surface.
Discussion
Here, we report the isolation and characterization of strain PA11, a food-grade derivative of the model probiotic L. rhamnosus GG that is known for its strong mucosal adherence due to the mucus-binding capacity of SpaC, which is the decorating and cap pilin of the SpaCBA pili (Kankainen et al. 2009). Using a combination of genomics and proteomics, immune EM visualization, and a set of functional analyses, we showed that strain PA11 has a mutation in house-keeping sortase A (srtA) gene, is deficient in mucus binding, and secretes its SpaCBA pili and other sortase-dependent proteins in the environment.
Sortase A is known to be involved in the covalent anchoring of the newly synthesized pili to lipid II structures in peptidoglycan (Marraffini et al. 2006; Clancy et al. 2010; Jacobitz et al. 2017). The mutation in the srtA gene of strain PA11 (nucleotide change at position 632 from G → A) affects amino acid residue 211 and results in an amino acid change from a positively charged Arg into a neutral Gln (Supplementary File 1, Fig. 2). Alignment of the L. rhamnosus GG sortase A (CAR88038.1) to other members of the highly conserved sortase A family cd06165, revealed that the affected Arg residue is part of the strongly conserved catalytic triad of the sortase A family: His, Cys (part of the TLXTC catalytic domain), and Arg (Fig. 2) (Marraffini et al. 2004). The conserved Arg residue at position 211 is essential for catalysis. It functions as a base, facilitating thiolate formation during sortase-mediated cleavage and transpeptidation, thus stabilizing the key substrate-enzyme intermediate required for efficient catalysis. Disruption of this Arg in Staphylococcus aureus severely reduced, but not completely eliminated, sortase A function, by almost completely abolishing transpeptidation (Marraffini et al. 2004). Given the strong conservation and essentiality of this Arg residue in SrtA efficacy (Fig. 2), its disruption explains the genotypic basis for the strain PA11 phenotype (Fig. 1). Indeed, strain PA11 is characterized by release of mature pili in the direct environment of the cells, with only a small fraction potentially still being attached to the cell surface (Fig. 1a). This is in accordance with reports in S. aureus (Marraffini et al. 2004) and might point towards the underlying mechanism of SrtA being unable to anchor the SpaCBA pili to lipid II. Abolishment of the housekeeping sortase SrtA has been shown to affect proper attachment of pili and by extension of other sortase-dependent proteins to the cell wall also in other Gram-positive bacteria, like Corynebacterium diptheriae (Swaminathan et al. 2007), Lactobacillus plantarum WCFS1 (Remus et al. 2013), Lactobacillus acidophilus NCFM, and Lactobacillus gasseri (Call et al. 2015). In these studies, it was reported that the failure to retain these sortase-dependent proteins to the cell surface often results in the loss of important strain phenotypes, like immunomodulation or adhesion (Remus et al. 2013; Call et al. 2015). These observations corroborate our findings further and support our conclusion that the mutation resulting in the Arg211Gln substitution in the housekeeping SrtA enzyme results in secretion of mature pili in the environment and ultimately in a strong reduced mucus-binding capacity of strain PA11. It will be interesting to validate this further using a site-directed mutagenesis approach in future studies although this may be technically challenging as L. rhamnosus GG is not so amenable to genetic changes as other model strains like Lactococcus lactis. Hence, in a previous work, we used Lactococcus lactis expressing L. rhamnosus GG pili for studying the effect of the sortase genes (both srtA and srtC1) and showed that removal or functional disruption of the housekeeping sortase resulted in a similar phenotype in this heterologous host (Douillard et al. 2014).
Microscopic analysis using pili-directed antibodies as a means to visualize SpaCBA pili and their components, resulted in the confirmation that PA11 secretes its pili in the environment, while only a small fraction of its pili remains bound to the cell envelope. Further Western blot analysis illustrated that these are low molecular weight pili and that the high molecular weight pilin agglomerates are secreted in the milieu surrounding PA11 cells. Retention of these low molecular weight pili could be caused by very low residual sortase A activity, but more likely options include the linkage of pili oligomers by hydrophobic interactions to the bacterial envelope or weak sortase C activity linking SpaB to lipid II, as has been earlier suggested (Douillard et al. 2014). Overall, the lack of high molecular weight pili in PA11 supports our finding that PA11 performs poorly in mucus-binding assays (with a more than 6-fold reduction) compared to its parental L. rhamnosus GG strain (wild type, Fig. 1d). This confirms that the SpaCBA pili are the main molecules involved in the high mucus adhesion phenotype of L. rhamnosus GG (Kankainen et al. 2009). The low residual mucus binding of strain PA11 may be due to the presence of some low molecular weight pili with the mucus-binding SpaC on its cell envelope, which is in accordance with our Immuno EM observations (Fig. 1). Apart from the SpaCBA pili, there are also other adhesins present on the cell surface of L. rhamnosus GG, which play a minor role in mucus binding, like the MBF protein, which is also a sortase-dependent protein (von Ossowski et al. 2011).
The use L. rhamnosus PA11 may have biotechnological advantages as it allows relative cost-effective and efficient enrichment of sortase-dependent proteins from the supernatant. This is extremely challenging to achieve using wild-type L. rhamnosus GG given the covalent association of its sortase-dependent proteins to the cell surface. The SpaCBA pili in particular have been implicated as the key modulators of L. rhamnosus GG extraordinary adhesion capacity, but also have a strong therapeutic potential. Earlier work has identified them as immunomodulatory molecules (Lebeer et al. 2012; Tytgat et al. 2016b), and they can be used in efforts to combat the important nosocomial pathogen Vancomycin-resistant enterococci (VRE) (Tytgat et al. 2016a).
In total, we found seven sortase-dependent proteins to be enriched in the secretome of PA11. Based on theoretical predictions, there is genomic potential of 19 sortase-dependent proteins present in L. rhamnosus GG (Douillard et al. 2014). However, we know from previous research that not all of these are actually expressed by L. rhamnosus strains, e.g., SpaFED pili (Reunanen et al. 2012). Interestingly, among the secreted sortase-dependent proteins in the PA11 strains are several proteins with therapeutic potential. The generated strain PA11 can thus be used to study the therapeutic potential of other sortase-dependent proteins with an immunomodulatory potential. These include the lactocepin protein (LGG_RS13070), a putative protease also known as PrtP, that is found in related Lactobacilli and in L. casei and that was shown to have potent immunomodulatory properties (von Schillde et al. 2012). Another abundant and large (2603 amino acids) secreted protein is the adhesion exoprotein (LGG_RS13990), a Leucin repeat protein that is partly conserved in Lactobacilli but presently has no assigned function and hence needs further functional characterization.
In conclusion, the here-characterized L. rhamnosus GG–derived PA11 strain opens novel avenues towards the exploration of these key molecules, both in a fundamental and therapeutic setting. Further enhancing the potential of strain PA11 is its non-GMO nature, rendering both the strain and its products safe for use in humans given the current regulations (EU 2001).
Data availability
The genome sequence of L. rhamnosus GG PA11 can be found in the NCBI Sequence Read Archive (SRA) database with accession number SAMN04440354, and all sequencing data was deposited as part of BioProject PRJNA309744. The proteomics data is included in supplementary material. L. rhamnosus PA11 has been deposited at the DSM-Z Braunschweig, Germany and is available under number DSM 111733.
References
Ardita CS, Mercante JW, Kwon YM, Luo L, Crawford ME, Powell DN, Jones RM, Neish AS (2014) Epithelial adhesion mediated by pilin SpaC is required for Lactobacillus rhamnosus GG-induced cellular responses. Appl Environ Microbiol 80(16):5068–5077. https://doi.org/10.1128/AEM.01039-14
Arvola T, Laiho K, Torkkeli S, Mykkanen H, Salminen S, Maunula L, Isolauri E (1999) Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: a randomized study. Pediatrics 104(5):e64. https://doi.org/10.1542/peds.104.5.e64
Avall-Jaaskelainen S, Lindholm A, Palva A (2003) Surface display of the receptor-binding region of the Lactobacillus brevis S-layer protein in Lactococcus lactis provides nonadhesive lactococci with the ability to adhere to intestinal epithelial cells. Appl Environ Microbiol 69(4):2230–2236. https://doi.org/10.1128/aem.69.4.2230-2236.2003
Call EK, Goh YJ, Selle K, Klaenhammer TR, O'Flaherty S (2015) Sortase-deficient lactobacilli: effect on immunomodulation and gut retention. Microbiology 161(Pt 2):311–321. https://doi.org/10.1099/mic.0.000007
Chandrasekharan B, Saeedi BJ, Alam A, Houser M, Srinivasan S, Tansey M, Jones R, Nusrat A, Neish AS (2019) Interactions between commensal Bacteria and enteric neurons, via FPR1 induction of ROS, increase gastrointestinal motility in mice. Gastroenterology 157(1):179–192 e172. https://doi.org/10.1053/j.gastro.2019.03.045
Clancy KW, Melvin JA, McCafferty DG (2010) Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Biopolymers 94(4):385–396. https://doi.org/10.1002/bip.21472
Douillard FP, Rasinkangas P, von Ossowski I, Reunanen J, Palva A, de Vos WM (2014) Functional identification of conserved residues involved in Lactobacillus rhamnosus strain GG sortase specificity and pilus biogenesis. J Biol Chem 289(22):15764–15775. https://doi.org/10.1074/jbc.M113.542332
Douillard FP, Rasinkangas P, Bhattacharjee A, Palva A, de Vos WM (2016) The N-terminal GYPSY motif is required for pilin-specific sortase SrtC1 functionality in Lactobacillus rhamnosus strain GG. PLoS One 11(4):e0153373. https://doi.org/10.1371/journal.pone.0153373
EU, T.E.P.a.t.C.o.t (2001) Directive 2001/18/EC of the European Parliament and of the council of 12 march 2001 on the deliberate release into the environment of genetically modified organisms and repealing council directive 30/2203EEC. OJ L106 (1)
Hatakka K, Savilahti E, Ponka A, Meurman JH, Poussa T, Nase L, Saxelin M, Korpela R (2001) Effect of long term consumption of probiotic milk on infections in children attending day care centres: double blind, randomised trial. BMJ 322(7298):1327. https://doi.org/10.1136/bmj.322.7298.1327
Hendrickx AP, Budzik JM, Oh SY, Schneewind O (2011) Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol 9(3):166–176. https://doi.org/10.1038/nrmicro2520
Jacobitz AW, Kattke MD, Wereszczynski J, Clubb RT (2017) Sortase transpeptidases: structural biology and catalytic mechanism. Adv Protein Chem Struct Biol 109:223–264. https://doi.org/10.1016/bs.apcsb.2017.04.008
Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM (2009) Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A 106(40):17193–17198. https://doi.org/10.1073/pnas.0908876106
Kant A, von Ossowski I, Palva A, Krishnan V (2016) Crystallization and X-ray crystallographic analysis of the adhesive SpaC pilin subunit in the SpaCBA pilus of gut-adapted Lactobacillus rhamnosus GG. Protein Pept Lett 23(4):365–371. https://doi.org/10.2174/0929866523666160106153055
Kumpu M, Kekkonen RA, Kautiainen H, Jarvenpaa S, Kristo A, Huovinen P, Pitkaranta A, Korpela R, Hatakka K (2012) Milk containing probiotic Lactobacillus rhamnosus GG and respiratory illness in children: a randomized, double-blind, placebo-controlled trial. Eur J Clin Nutr 66(9):1020–1023. https://doi.org/10.1038/ejcn.2012.62
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5(2):R12. https://doi.org/10.1186/gb-2004-5-2-r12
Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, de Vos WM, De Keersmaecker SC, Vanderleyden J (2012) Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol 78(1):185–193. https://doi.org/10.1128/AEM.06192-11
Lee CL, Hsiao HH, Lin CW, Wu SP, Huang SY, Wu CY, Wang AH, Khoo KH (2003) Strategic shotgun proteomics approach for efficient construction of an expression map of targeted protein families in hepatoma cell lines. Proteomics 3(12):2472–2486. https://doi.org/10.1002/pmic.200300586
Luoto R, Ruuskanen O, Waris M, Kalliomaki M, Salminen S, Isolauri E (2014) Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: a randomized, placebo-controlled trial. J Allergy Clin Immunol 133(2):405–413. https://doi.org/10.1016/j.jaci.2013.08.020
Mandlik A, Swierczynski A, Das A, Ton-That H (2008) Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol 16(1):33–40. https://doi.org/10.1016/j.tim.2007.10.010
Marraffini LA, Ton-That H, Zong Y, Narayana SV, Schneewind O (2004) Anchoring of surface proteins to the cell wall of Staphylococcus aureus. A conserved arginine residue is required for efficient catalysis of sortase a. J Biol Chem 279(36):37763–37770. https://doi.org/10.1074/jbc.M405282200
Marraffini LA, Dedent AC, Schneewind O (2006) Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev 70(1):192–221. https://doi.org/10.1128/MMBR.70.1.192-221.2006
Nishiyama K, Ueno S, Sugiyama M, Yamamoto Y, Mukai T (2016) Lactobacillus rhamnosus GG SpaC pilin subunit binds to the carbohydrate moieties of intestinal glycoconjugates. Anim Sci J 87(6):809–815. https://doi.org/10.1111/asj.12491
Parekh S, Vinci VA, Strobel RJ (2000) Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol 54(3):287–301. https://doi.org/10.1007/s002530000403
Rasinkangas P, Reunanen J, Douillard FP, Ritari J, Uotinen V, Palva A, de Vos WM (2014) Genomic characterization of non-mucus-adherent derivatives of Lactobacillus rhamnosus GG reveals genes affecting pilus biogenesis. Appl Environ Microbiol 80(22):7001–7009. https://doi.org/10.1128/AEM.02006-14
Remus DM, Bongers RS, Meijerink M, Fusetti F, Poolman B, de Vos P, Wells JM, Kleerebezem M, Bron PA (2013) Impact of Lactobacillus plantarum sortase on target protein sorting, gastrointestinal persistence, and host immune response modulation. J Bacteriol 195(3):502–509. https://doi.org/10.1128/JB.01321-12
Reunanen J, von Ossowski I, Hendrickx AP, Palva A, de Vos WM (2012) Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol 78(7):2337–2344. https://doi.org/10.1128/AEM.07047-11
Saxelin M, Tynkkynen S, Mattila-Sandholm T, de Vos WM (2005) Probiotic and other functional microbes: from markets to mechanisms. Curr Opin Biotechnol 16(2):204–211. https://doi.org/10.1016/j.copbio.2005.02.003
Sega GA (1984) A review of the genetic effects of ethyl methanesulfonate. Mutat Res 134(2–3):113–142. https://doi.org/10.1016/0165-1110(84)90007-1
Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H (2007) Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol 66(4):961–974. https://doi.org/10.1111/j.1365-2958.2007.05968.x
Szajewska H, Kolodziej M (2015) Systematic review with meta-analysis: Lactobacillus rhamnosus GG in the prevention of antibiotic-associated diarrhoea in children and adults. Aliment Pharmacol Ther 42(10):1149–1157. https://doi.org/10.1111/apt.13404
Tripathi P, Beaussart A, Alsteens D, Dupres V, Claes I, von Ossowski I, de Vos WM, Palva A, Lebeer S, Vanderleyden J, Dufrene YF (2013) Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano 7(4):3685–3697. https://doi.org/10.1021/nn400705u
Tytgat HL, Douillard FP, Reunanen J, Rasinkangas P, Hendrickx AP, Laine PK, Paulin L, Satokari R, de Vos WM (2016a) Lactobacillus rhamnosus GG outcompetes Enterococcus faecium via mucus-binding pili: evidence for a novel and heterospecific probiotic mechanism. Appl Environ Microbiol 82(19):5756–5762. https://doi.org/10.1128/AEM.01243-16
Tytgat HL, van Teijlingen NH, Sullan RM, Douillard FP, Rasinkangas P, Messing M, Reunanen J, Satokari R, Vanderleyden J, Dufrene YF, Geijtenbeek TB, de Vos WM, Lebeer S (2016b) Probiotic gut microbiota isolate interacts with dendritic cells via glycosylated heterotrimeric pili. PLoS One 11(3):e0151824. https://doi.org/10.1371/journal.pone.0151824
Vesterlund S, Paltta J, Karp M, Ouwehand AC (2005) Measurement of bacterial adhesion-in vitro evaluation of different methods. J Microbiol Methods 60(2):225–233. https://doi.org/10.1016/j.mimet.2004.09.013
von Ossowski I, Reunanen J, Satokari R, Vesterlund S, Kankainen M, Huhtinen H, Tynkkynen S, Salminen S, de Vos WM, Palva A (2010) Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl Environ Microbiol 76(7):2049–2057. https://doi.org/10.1128/AEM.01958-09
von Ossowski I, Satokari R, Reunanen J, Lebeer S, De Keersmaecker SC, Vanderleyden J, de Vos WM, Palva A (2011) Functional characterization of a mucus-specific LPXTG surface adhesin from probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol 77:4465–4472. https://doi.org/10.1128/AEM.02497-10
von Schillde MA, Hormannsperger G, Weiher M, Alpert CA, Hahne H, Bauerl C, van Huynegem K, Steidler L, Hrncir T, Perez-Martinez G, Kuster B, Haller D (2012) Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 11(4):387–396. https://doi.org/10.1016/j.chom.2012.02.006
Wolvers D, Antoine JM, Myllyluoma E, Schrezenmeir J, Szajewska H, Rijkers GT (2010) Guidance for substantiating the evidence for beneficial effects of probiotics: prevention and management of infections by probiotics. J Nutr 140(3):698S–712S. https://doi.org/10.3945/jn.109.113753
Zheng J, Wittouck S, Salvetti E, Franz C, Harris HMB, Mattarelli P, O'Toole PW, Pot B, Vandamme P, Walter J, Watanabe K, Wuyts S, Felis GE, Ganzle MG, Lebeer S (2020) A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70:2782–2858. https://doi.org/10.1099/ijsem.0.004107
Acknowledgments
We are grateful to Julian Ket (Wageningen University) for experimental assistance in analysis of the secretome and thank Prof. Markus Aebi for supporting the MS analysis (ETH Zurich). We gratefully acknowledge Dr. Ingemar von Ossowski of University of Helsinki for providing the antibodies against SpaA and SpaC pilins. Additionally, we are grateful to the personnel of Sequencing and Electron Microscopy Units of the Institute of Biotechnology, University of Helsinki.
Funding
This project, including the efforts of PR, FPD, and JRitari were funded by the European Research Council (grant 250172, Microbes Inside to WMdV) and Academy of Finland (grants 252123 for FPD, 299749 for JReunanen, and 137389 for WMdV). The work of HPLT and WMdV was partly funded by the SIAM Gravity Grant 024.002.002 and Spinoza Award of the Netherlands Organization for Scientific Research of WMdV.
Author information
Authors and Affiliations
Contributions
PR, FD, AP, and WMdV contributed to conception and design of the study; PR and HT carried out experimental procedures; JReunanen performed immuno-EM analysis; JRitari was responsible for Illumina sequence processing of all strains; SA and C-WL assisted with proteomics; and HT and PR wrote the manuscript and made the figures. All authors contributed to manuscript revision and read and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval, consent to participate, and consent for publication
Not applicable.
Code availability
Not applicable
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary File 1
SNP tables from shotgun and single-molecule genome sequencing of PA11 (XLSX 33 kb)
Supplementary File 2
Proteomics of the secreted protein fraction of the PA11 strain compared to wild-type L. rhamnosus GG (XLSX 71 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tytgat, H.L.P., Rasinkangas, P., Ritari, J. et al. Selection and characterization of a SpaCBA pilus-secreting food-grade derivative of Lacticaseibacillus rhamnosus GG. Appl Microbiol Biotechnol 105, 1123–1131 (2021). https://doi.org/10.1007/s00253-020-11051-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-11051-7