Abstract
Schistosome infection is recognized as a potentially modifiable risk factor for HIV in women by the World Health Organization. Alterations in cervicovaginal bacteria have been associated with HIV acquisition and have not been studied in schistosome infection. We collected cervical swabs from Tanzanian women with and without S. mansoni and S. haematobium to determine effects on cervicovaginal microbiota. Infected women were treated, and follow-up swabs were collected after 3 months. 16S rRNA sequencing was performed on DNA extracted from swabs. We compared 39 women with S. mansoni with 52 uninfected controls, and 16 with S. haematobium with 27 controls. S. mansoni-infected women had increased abundance of Peptostreptococcus (p = 0.026) and presence of Prevotella timonesis (p = 0.048) compared to controls. High-intensity S. haematobium infection was associated with more diverse cervicovaginal bacterial communities than uninfected controls (p = 0.0159). High-intensity S. mansoni infection showed a similar trend (p = 0.154). At follow-up, we observed increased alpha diversity in S. mansoni (2.53 vs. 1.72, p = 0.022) and S. haematobium (2.05 vs. 1.12, p = 0.066) infection groups compared to controls. Modifications in cervicovaginal microbiota, particularly increased diversity and abundance of taxa associated with bacterial vaginosis and HIV (Peptostreptococcus, Prevotella), were associated with schistosome infection.
Similar content being viewed by others
Introduction
Increased prevalence and incidence of HIV in African women with schistosome infections is supported by studies of both S. haematobium [1,2,3], which predominantly affects the urogenital tract, and S. mansoni [4, 5], which concentrates in the gastrointestinal tract. One longitudinal study reported that S. mansoni-infected women had a subsequent 2.8-fold increased risk for HIV acquisition [5]. Another noted a 1.4-fold increased hazard of HIV acquisition in women with S. haematobium infection, and a 1.3-fold increased hazard in women with S. mansoni infection that did not reach significance (p = 0.12) [6]. Though not all studies have confirmed these associations [7, 8], schistosome infection has been declared a potentially modifiable risk factor for HIV acquisition by both the World Health Organization and the Joint United Nations Program on HIV/AIDS [9, 10].
The mechanism for increased risk of HIV acquisition in schistosome-infected women is not fully understood. Increased risk in schistosome-infected women but not men [1, 5, 6, 11], suggests that increased HIV susceptibility may be attributable to genital mucosal effects of schistosome infection. Previously, we showed that women with S. haematobium had altered cervical mucosal gene expression and lower cervicovaginal interleukin-15 levels compared to schistosome-uninfected women, whereas women with S. mansoni did not have these alterations [12]. Possible genital tract alterations by which S. mansoni may increase risk for HIV acquisition remain unclear.
Increased cervicovaginal bacterial diversity is a putative risk factor for HIV acquisition in African women [13,14,15] and to our knowledge has not been investigated in schistosome-infected women. There are, however, schistosome-associated alterations in gut and urinary bacterial communities. S. mansoni infection has been associated with lower intra- individual gut bacterial diversity in mice and children [16, 17], and S. haematobium infection is associated with gut microbiota suggestive of dysbiosis [18, 19] and with immune-stimulatory urinary bacterial taxa [20]. Therefore, it is plausible that schistosome infections may affect cervicovaginal bacterial communities given that parasite eggs concentrate in the genital tract in S. haematobium, and to a lesser extent in S. mansoni [21].
Our objective was to investigate the effects of S. haematobium, S. mansoni and praziquantel treatment on cervicovaginal bacterial communities. We hypothesized that schistosome-infected women would have altered cervicovaginal microbiota compared to uninfected women, and that these alterations would vary by schistosome species and after anti-schistosome treatment. We therefore compared women with and without S. haematobium and women with and without S. mansoni separately. In addition, we compared microbiota of women who were infected with S. haematobium or S. mansoni, treated with praziquantel, and confirmed to be schistosome-uninfected at follow-up. A deeper understanding of the relationship between schistosome infections and cervicovaginal bacterial communities could suggest additional treatment strategies by which the sequelae of schistosome infections in women could be managed, with the ultimate goal of decreasing morbidity and preventing HIV infections in this vulnerable population.
Methods
Ethics
All participants provided written informed consent, which was obtained in private by a nurse in Kiswahili. Those with schistosome and other genital tract infections received free treatment for themselves and their partners if indicated. Women with newly diagnosed HIV were referred to the nearest HIV treatment center for free ongoing care.
Ethical approval for this study was provided by the Joint Research Ethics Committee of Bugando Medical Centre/the Catholic University for Health and Allied Sciences in Mwanza, Tanzania (CREC/171/2017), the National Institute for Medical Research (NIMR) in Dar es Salaam (NIMR/HQ/R.8a/Vol.IX/2446), and Weill Cornell Medicine in New York (1612017800).
Eligibility screening
We invited reproductive-aged women living in rural Tanzanian villages with high prevalence of S. haematobium or S. mansoni infection to undergo screening for HIV and schistosome infections, as previously described [12, 22]. Women from S. mansoni-endemic villages were screened between July 2017 and January 2018 and women from S. haematobium-endemic villages were screened between June and September 2018. S. mansoni- and S. haematobium-endemic villages were >60 km apart. In this community-based sample, women were apparently healthy and had no obvious clinical conditions. HIV counseling and testing was performed by a trained nurse using the Determine HIV-1/2 test (Alere, MA), with positive results confirmed by Uni-Gold HIV-1/2 test (Trinity BioTech, Ireland), per Tanzanian national guidelines. Urine samples were collected, filtered with a single syringe (two filtrations of 10 mL each to increase sensitivity), and examined microscopically for S. haematobium ova by trained parasitologists. Five slides per stool sample [23] were examined for S. mansoni ova by Kato-Katz technique. Serum was stored at −20 °C within 10 h of collection and schistosome circulating anodic antigen (CAA) was quantified at the NIMR laboratory in Mwanza using up-converting phosphor technology with CAA values >30 pg/mL considered positive [4, 24, 25].
Study procedures
Women received a follow-up card to return for results in 1–2 weeks. Each received schistosome test results privately, with free praziquantel treatment if infected. Women confirmed positive for S. haematobium or S. mansoni by both microscopy and CAA, or those confirmed to be negative both by microscopy and CAA, were eligible for cohort participation. For every schistosome-infected woman enrolled, the following uninfected woman from her village was also invited to participate in the cohort. Thus, infected and uninfected women were recruited from the same schistosome-endemic villages. Pregnant women were screened but not eligible for cohort participation because endocervical brushing would be performed.
Cohort-eligible women participated in a structured private interview a nurse in Kiswahili, followed by gynecologic examination with specimen collection. For this study, a sealed Copan swab was touched straight to the cervical face and vaginal fornix, and then placed immediately back into the transport container, which was resealed and placed on ice (ThermoScientificTM COPAN ESwabTM, U.S.). All women were tested for trichomoniasis, chlamydia and gonorrhea and screened for cervical cancer using acetic acid, as per Tanzanian national guidelines, with follow-up referrals provided to Bugando Medical Centre for free ongoing cervical cancer screening and care as needed [26]. Women who tested positive for trichomoniasis were provided same-day treatment free of charge and women positive for gonorrhea or chlamydia were provided free treatment several weeks later, after PCR testing in the central laboratory.
Participants provided 1–2 phone numbers to allow study nurses to maintain contact with them for the study’s duration. Participants returned after 3 months, at the midpoint in their menstrual cycles. At follow-up, women provided urine, blood, stool, and gynecologic samples, and were again tested for STIs and treated as indicated. Women who tested positive for schistosomiasis by urine, stool, or serum at any follow-up received directly observed praziquantel treatment.
Sample processing and testing
Point-of-care Trichomonas testing was performed during the gynecologic examination (Osom Rapid Test, Sekisui Diagnostics, U.S.). For chlamydia and gonorrhea testing at NIMR, DNA was extracted from cervicovaginal swab specimens by QIAamp DNA mini-kit (Qiagen, Germany) for quantitative polymerase chain reaction analysis using the Artus CT/NG RGQ kit.
Cervical swabs were placed on ice and stored at −80 °C within four hours of collection at the NIMR laboratory until shipment to Weill Cornell Medicine on dry ice.
16S rRNA sequencing on vaginal swab
Minor adaptions were made to the previously described phenol/chloroform/isoamyl extraction for vaginal swabs [27]. After thawing to room temperature, 500 µl of extraction buffer (200 mM Tris, pH 8.0, 200 mM NaCl, and 20 mM EDTA) was added to the tube containing the swab and vortexed to ensure sufficient binding. Liquid containing DNA extraction buffer and sample was then suspended in 200 μl of 20% SDS, 500 µl of phenol/chloroform/isoamyl alcohol (24:24:1), and 500 µl of 0.1-mm-diam zirconia/silica beads (BioSpec Products, US). Cells were lysed by mechanical disruption by bead beater (FisherScientific, U.S.) for 2 min. Two rounds of phenol/chloroform/isoamyl alcohol extraction were performed. DNA was precipitated with 40 μl sodium acetate and 880 μL ethanol, incubated at −80 °C for 20 min, and pelleted at max speed for 20 min. The supernatant was removed and pellet resuspended in 200 µl TE buffer with 100 µg/ml RNase (ThermoScientific). Isolated DNA was further purified with QIAamp Mini Spin Columns (Qiagen).
The V4-V5 region of the 16S rRNA gene was amplified using primers 563F (5′-nnnnnnnn-NNNNNNNNNNNN-AYTGGGYDTAAAGNG-3′) and 926R (5′-nnnnnnnn-NNNNNNNNNNNN-CCGTCAATTYHTTTRAGT-3′) as previously described, with extraction blanks serving as controls [28]. Amplicons were purified using the Qiaquick PCR Purification kit (Qiagen) and quantified with the Agilent 2200 tape station (Agilent Technologies, Germany). PCR products were then pooled at equimolar amounts. Libraries were prepared with TruSeq DNA library preparation kit (Illumina, US), per the manufacturer’s instructions, and sequenced on the Illumina MiSeq platform using a paired-end 250 × 250-bp kit.
16S rRNA phylogenetic analysis
Paired 16S rRNA (V4-V5) end reads were merged and demultiplexed. The DADA2 pipeline [29] was used for maximum expected error filtering (Emax = 10) [30], with amplicon sequence variant (ASV) grouping (by unique 16S sequence). Singleton sequences were removed. Nucleotide BLAST of representative sequences from each ASV was used for taxonomic classification, with NCBI RefSeq as the reference database. Minimum threshold for E values was 1e−10. A de novo neighbor-joining phylogenetic tree of 16S ASV sequences was constructed using MUSCLE [29].
Statistical analysis
We calculated that enrolling 15 women with schistosome infection and 25 without would provide >86% power to detect a difference in alpha diversity of 1.4 versus 1.8, assuming a standard deviation of 0.4 [30].
For univariate analysis to identify differently abundant and present taxa in S. mansoni-infected vs. uninfected women from S. mansoni-endemic villages and S. haematobium-infected vs. uninfected women from S. haematobium-endemic villages, we first filtered out extremely low prevalent taxa from downstream analysis in order to reduce problems induced by spurious operational taxonomic units (OTUs) [31, 32]. In order to deal with excessive zeros in the data, we then classified taxa as either prevalent or less prevalent and formulated statistical tasks adaptively to the prevalence of taxa. There was a natural separation of taxa clusters based on the prevalence with a cutoff near one third [33]. Therefore, for prevalent taxa, defined as taxa present in at least one third (≥32.6%) of women, we focus on identifying taxa that show differential abundance by infection status using rank-sum tests. For less prevalent taxa, actual composition is largely determined by zero counts. Instead of comparing small and zero abundance in most cases, we aim to detect presence/absence of taxa patterns [34] that were associated with infection status using logistic regression (or Fisher’s Exact test if any cell count was <5). As per multiple previous studies [13, 14, 35], we did not correct for multiple statistical tests when comparing bacterial taxa between infection groups. Instead, we used follow-up data to validate baseline findings.
Alpha diversity was calculated using the Shannon index measuring within-sample bacterial community diversity and compared using rank-sum test by infection intensity. Beta diversity was measured using principal coordinate analysis with the Bray-Curtis distance metric [36, 37]. Infection intensity was categorized as low (CAA 30-3,000 pg/mL) and high (CAA > 3,000 pg/mL) [38]. Analyses were performed using R version 3.6.1.
Results
In S. mansoni-endemic villages, 41 women with S. mansoni, one with S. haematobium-S. mansoni coinfection, and 58 uninfected women consented to cohort participation. The woman with S. haematobium–S. mansoni coinfection was excluded from analysis. Sequencing data were successfully obtained from 39 S. mansoni-infected (22 with low-intensity and 17 with high-intensity infection) and 53 uninfected cervical swabs at baseline; the remaining seven samples did not amplify after DNA extraction.
In S. haematobium-endemic villages, 47 women consented to cohort participation. Of these, 18 had S. haematobium infection and 29 were uninfected at enrollment. Sequencing data were successfully obtained from 17 S. haematobium-infected (12 with low-intensity and five with high-intensity infection) and 27 uninfected baseline cervical swabs; three samples did not amplify after DNA extraction and one did not generate sufficient sequencing depth.
Meta data, OTUs, taxa, and phylogenetic trees are included for S. mansoni and S. haematobium baseline and follow-up in Supplementary Data Files 1–16.
Overall bacterial composition
Lactobacillus was the most common taxa at genus level across all samples, with a mean relative abundance of 0.28, followed by Gardnerella, Megasphaera, and Sneathia. In addition, 27.2% (37/136) of samples had Lactobacillus encompassing >0.5 of their relative abundance. Mean alpha diversity across all samples was 1.94.
S. mansoni baseline
S. mansoni-infected women had similar numbers of sexual partners in the last year, rates of HIV infection, frequencies of vaginal cleansing, and rates of current sexually transmitted infections (STIs) compared to schistosome-uninfected women from S. mansoni-endemic villages (Table 1). Women with S. mansoni more frequently experienced food suffrage (71.8% vs. 42.3%) and were slightly younger at first sexual encounter (17 vs. 17.5 years) (p < 0.05 for all).
Neither beta diversity (p = 0.515, Fig. 1a, Supplementary Fig. 1a) nor alpha diversity differed between infected and uninfected women at baseline (2.03 vs. 1.88, p = 0.320, Fig. 2a). Those with high-intensity S. mansoni infection tended towards increased alpha diversity compared to uninfected women (2.04 vs. 1.88, p = 0.154, Fig. 2a).
Principal coordinate analysis using Bray-Curtis distance metric on schistosome infected and uninfected women at baseline and 3-month follow-up. Women infected with S. haematobium at 3-month follow-up (d) are more clustered compared to uninfected women (c), or to women with S. mansoni infection at baseline (a) or 3-month follow-up (b). This indicates that infected women have cervicovaginal microbiomes that are more similar to each other than uninfected women.
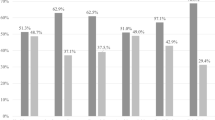
Differences in alpha diversity, as shown by Shannon Diversity Index, between uninfected, low intensity infected, and high intensity infected women at baseline and 3-month follow-up from S. mansoni and S. haematobium-endemic villages. The plots display medians (dark horizontal bars) and interquartile ranges (boxes), with error bars representing 1.5 times the interquartile range or minimum/maximum values. Women with high-intensity S. mansoni infection had a trend towards increased diversity compared to uninfected women at baseline (p = 0.154). Women with high-intensity S. haematobium infection had significantly increased diversity compared to their uninfected counterparts at baseline (p = 0.016). Women with low-intensity infection S. mansoni or S. haematobium infection had increased diversity compared to their uninfected counterparts at follow-up (p = 0.022 and p = 0.066).
Overall microbial composition of S. mansoni-infected and uninfected women is shown at the genus level (Fig. 3). Relative abundance and presence/absence of bacteria at different phylogenetic levels were compared between infected and uninfected women from S. mansoni-endemic villages at baseline. Prevotella timonensis was more often detected in S. mansoni-infected than uninfected women (p = 0.048). Peptostreptococcus anaerobius was more abundant in S. mansoni-infected women (p = 0.040), at both genus and family levels (Fig. 4). Peptoniphilaceae, Tissierellales, and Tissierella were more abundant in infected women at the family, order and class levels (p = 0.046, p = 0.046, p = 0.045, respectively). Lactobacillus and Lactobacillus iners were both reduced in women with high-intensity S. mansoni infection compared to uninfected and low intensity infected women, though this did not reach significance (p = 0.273 and p = 0.141, respectively).
Relative abundance of the 11 most abundant cervicovaginal microbiota at the genus level for uninfected, low intensity infected, and high intensity infected women with S. mansoni (upper horizontal band) and S. haematobium (lower horizontal band) at baseline. Each vertical bar represents a successfully sequenced cervicovaginal sample. Labels above each bar indicate infection status: “U” for uninfected, “L” for low-intensity infection, and “H” for high-intensity infection. Women with S. mansoni infection had increased Peptostreptococcus (shown in dark blue, p = 0.026) and women with high-intensity S. mansoni infection showed a trend toward decreased Lactobacillus (p = 0.273). No significant differences were observed in individual taxa between S. haematobium infected and uninfected women.
Infected and uninfected at baseline and 3-month follow-up. Differences in relative abundance of Peptostreptococcus anaerobius between S. mansoni infected and uninfected women at baseline and 3-month follow-up. The plots display medians (dark horizontal bars) and interquartile ranges (boxes), with error bars representing 1.5 times the interquartile range or maximum values. Women with S. mansoni infected had increased abundance of Peptostreptococcus anaerobius at baseline and at follow-up compared to uninfected women (p = 0.040 and p = 0.119, respectively).
S. mansoni follow-up
Sequencing data were successfully obtained from 59 women from S. mansoni-endemic villages at 3-month follow-up. Of the 34 women uninfected at baseline, 33 remained uninfected at follow-up, while one had acquired S. mansoni infection. Of the 25 women infected with S. mansoni at baseline, 18 were cured with praziquantel treatment, while seven remained infected. Therefore, sequencing data from 8 S. mansoni-infected and 51 uninfected women were used in follow-up analysis.
Women infected with S. mansoni at follow-up had significantly higher alpha diversity compared to uninfected women (2.53 vs. 1.72, p = 0.022, Fig. 2b). Of note, all infected women at follow-up had low-intensity infections. There was no significant difference in beta diversity between infected and uninfected women at follow-up (p = 0.359, Fig. 1b, Supplemental Fig. 1b). Differences in diversity at the genus level between infected and uninfected groups are shown in Fig. 5a.
Relative abundance of cervicovaginal microbiota at the genus level for uninfected, low intensity infected, and high intensity infected women with S. mansoni (upper band) and S. haematobium (lower band) at 3-month follow-up. Each vertical bar represents a successfully sequenced cervicovaginal sample. Labels above each bar indicate infection status: “U” for uninfected and “L” for low-intensity infection. Women with S. mansoni infection had increased Peptostreptococcus (as shown in dark blue, p = 0.049) and increased alpha diversity (p = 0.022). Women infected with S. haematobium infected also had increased alpha diversity (p = 0.017).
Taxa that were significantly different at baseline were tested at follow-up for validation of baseline findings. Peptostreptococcus was more abundant among infected women at the genus level (p = 0.049) and trended towards higher abundance at the family level (p = 0.063) (Fig. 4). Prevotella timonesis was more often detected in women infected at follow-up (p = 0.008). At the family, order, and class levels, Peptoniphilaceae, Tissierellales, and Tissierella were more abundant in the infected group (p = 0.042 for all), consistent with baseline findings.
S. haematobium baseline
S. haematobium-infected women had similar numbers sexual partners in the last year, and practices of vaginal cleansing compared to uninfected women from S. haematobium-endemic villages (Table 1).
Baseline alpha diversity (1.83 vs. 1.36, p = 0.390, Fig. 2c) and beta diversity (p = 0.604, Fig. 1c, Supplemental Fig. 1c) were not different. Women with high-intensity S. haematobium infections had significantly higher alpha diversity than uninfected women (2.43 vs. 1.37, p = 0.016) and than women with low-intensity S. haematobium infections (2.43 vs. 1.83, p = 0.038, Fig. 2c).
Overall microbial composition of S. haematobium-infected and uninfected women is shown at the genus level in Fig. 3b. In contrast to S. mansoni findings, at all phylogenetic levels, the relative abundance and presence or absence of bacteria were not significantly different between S. haematobium-infected and uninfected women at baseline. Women with high-intensity S. haematobium infection had reduced abundance of Lactobacillus iners compared to women with low-intensity infection, though this did not reach significance (p = 0.393).
S. haematobium follow-up
Sequencing data were successfully obtained from 34 women from S. haematobium-endemic villages at follow-up. Of the 20 women uninfected at baseline, 16 remained uninfected at follow-up, while four had acquired S. haematobium. Of the 12 women infected with S. haematobium at baseline, nine were cured with praziquantel treatment, while three remained infected. Therefore, sequencing data from seven S. haematobium-infected and 27 uninfected women were used in follow-up analysis.
Women with S. haematobium infection at follow-up showed a trend towards higher alpha diversity than uninfected women (2.05 vs. 1.12, p = 0.066, Fig. 2d). Of note, all infected women at follow-up had low-intensity infections. Women with S. haematobium had significantly different beta diversity than uninfected women, demonstrating that women with S. haematobium had microbiota more similar to each other than uninfected women at follow-up (p = 0.017, Fig. 5d, Supplementary Fig. 1d).
Discussion
Our novel study investigated the effects of schistosome infections on the cervicovaginal bacterial communities of African women to explore whether differences in microbiota could be a plausible and potentially modifiable mechanism by which schistosome infection may increase risk of HIV. We report that women with high-intensity S. mansoni or S. haematobium infections had increased cervicovaginal bacterial diversity compared to controls. S. mansoni and S. haematobium-infected women who had become infected or remained infected at follow-up also had higher diversity than uninfected women. In addition, women with S. mansoni infection, but unexpectedly not S. haematobium infection, had modified abundance and presence of specific bacterial taxa linked with increased vaginal pH compared to uninfected women.
Existing literature suggests that high-diversity, low Lactobacillus cervicovaginal microbiota are associated with increased risk of HIV acquisition [13, 39], and that African women have lower Lactobacillus and higher diversity than white European women [13, 14]. We report that women with high-intensity schistosome infection have increased cervicovaginal bacterial diversity compared to uninfected women. We also report a trend toward decreased Lactobacillus species and Lactobacillus iners in high-intensity S. mansoni infection, which did not reach significance likely because of sample size limitations. Both these results indicate that those with high-intensity schistosome infection may be at increased risk for acquiring HIV and align with our prior finding that women with high-intensity schistosome infection had a higher prevalence of HIV than women with low-intensity or without infections [4]. Our findings concur with studies reporting Lactobacillus dominance in <30% of African women, compared to 90% of European women [13].
Our findings also indicate that Peptostreptococcus anaerobius and Prevotella timonesis were more abundant and present, respectively, among S. mansoni infected compared to uninfected women. Peptostreptococcus and Prevotella are associated with increased vaginal pH [40] and bacterial vaginosis [41], both of which have been associated with increased HIV acquisition [41,42,43,44,45]. Thus, increased presence and abundance of Peptostreptococcus and Prevotella species could either increase or result from high vaginal pH and thereby contribute to increased HIV acquisition in S. mansoni-infected women. Prevotella has also been associated with genital inflammation and HIV acquisition [13], suggesting that increased Prevotella among S. mansoni-infected women could either contribute to or reflect increased mucosal inflammation in the genital tract.
Despite the the presence of eggs in the urogenital tract, S. haematobium-infected women had no significant differences in cervicovaginal microbiota compared to uninfected women. This contrast with S. mansoni-infected women could be due to distinct species differences, leading to different pathogenic and immunogenic effects in the host and different impacts of host immunity on the parasites. S. mansoni female worms produce approximately 10-fold more eggs than S. haematobium worms and maintain high egg production regardless of host age, while S. haematobium egg production decreases in older hosts [46,47,48]. Human autopsy studies have demonstrated large numbers of calcified S. haematobium eggs lodged in host tissues, while lower numbers of retained S. mansoni eggs suggests more efficient egg excretion [47]. S. mansoni egg excretion is facilitated by the parasite’s exploitation of vascular Peyer’s Patch gut lymphoid tissue, which enhances transit of eggs to the intestinal lumen while also damaging Peyer’s Patch structure and lymphoid cellularity [49]. An analogous lymphoid tissue does not exist for S. haematobium eggs migrating to the lumen of the bladder and may have differential systemic immune ramifications. In addition, both Peptostreptococcus and Prevotella are commonly found not only in the gut but also in the vagina, particularly in women with anaerobe-dominated cervicovaginal microbiota, and effects of S. mansoni could be more easily enhanced in the vaginal tract in which these populations are already present [13, 39]. Together, this body of data suggests multiple differential effects of S. mansoni on host tissue and host immunity, which may explain why it altered cervicovaginal microbiota while S. haematobium did not.
While ours is the first to report alterations in cervicovaginal microbial communities in women with and without schistosome infections, previous studies documented increased Prevotella in the gut microbiota of children and adolescents with S. haematobium infection [18, 19]. Studies investigating the relationship between S. mansoni and gut microbiota in mice [16] and children [17] reported subtle differences between infected and uninfected groups, though expectedly these differences do not match differences identified in cervicovaginal microbiota. Both these studies also found decreased alpha diversity in S. mansoni-infected individuals, consistent with gut dysbiosis and inflammation.
In contrast, increased diversity is associated with inflammation in cervicovaginal microbiota [13, 14] and women who were persistently schistosome-infected at follow-up had higher diversity. We hypothesize that this may be explained by immune changes caused by schistosome reinfection. Because praziquantel treatment was observed, women who were infected at baseline were likely cured and re-infected, or had juvenile worms that matured, before follow-up. The first six weeks of schistosome infection are characterized by a dynamic Th1-predominant dominant immune environment, which switches to Th2 predominance when egg-laying begins [50]. We hypothesize that this immune shift could affect microbial diversity, causing women who have been recently infected or re-infected to have increased diversity compared to those with chronic schistosome infections.
Our study was limited by small sample size, precluding our ability to control for differences between infected and uninfected groups and our ability to conduct a paired analysis, given that some baseline and follow-up sample pairs did not amplify. Multiple papers report that cervicovaginal bacterial communities are not associated with STIs, hormonal contraceptive use, or sexual behavior [13, 14]. In addition, women with schistosome infection in our study were more likely to experience food insecurity, which may have affected our findings. Further, the aforementioned potential confounders were similar between S. mansoni infected and uninfected groups and S. haematobium infected and uninfected groups, so these factors likely did not skew our results. Other limitations include that we did not determine vaginal pH and we could not examine the role of schistosome infection stage at baseline.
In conclusion, we have demonstrated longitudinally that S. mansoni and S. haematobium infections differentially modify cervicovaginal microbiota, and that both intensity of infection and stage of infection could affect these modifications. Changes observed in S. mansoni-infected women, including increased Peptostreptococcus and Prevotella timonesis, offer potential mechanisms for increased HIV incidence in S. mansoni-infected women. Observations of increased bacterial diversity in high-intensity infected and potentially newly-infected women with either schistosome species suggest that women with intense or recent schistosome infections may be more susceptible to HIV. These data warrant future studies to investigate the role of schistosome infection stage on cervicovaginal microbiota, as well as the potential to decrease HIV risk by normalizing altered cervicovaginal microbial communities in schistosome-infected women.
References
Kjetland EF, Ndhlovu PD, Gomo E, Mduluza T, Midzi N, Gwanzura L, et al. Association between genital schistosomiasis and HIV in rural Zimbabwean women. AIDS. 2006;20:593–600.
Downs JA, Mguta C, Kaatano GM, Mitchell KB, Bang H, Simplice H, et al. Urogenital schistosomiasis in women of reproductive age in Tanzania’s Lake Victoria region. Am J Trop Med Hyg. 2011;84:364–9.
Brodish PH, Singh K. Association between schistosoma haematobium exposure and human immunodeficiency virus infection among females in Mozambique. Am J Trop Med Hyg. 2016;94:1040–4.
Downs JA, van Dam GJ, Changalucha JM, Corstjens PLAM, Peck RN, de Dood CJ, et al. Association of Schistosomiasis and HIV infection in Tanzania. Am J Trop Med Hyg. 2012;87:868–73.
Downs JA, Dupnik KM, van Dam GJ, Urassa M, Lutonja P, Kornelis D, et al. Effects of schistosomiasis on susceptibility to HIV-1 infection and HIV-1 viral load at HIV-1 seroconversion: a nested case-control study. PLoS Negl Trop Dis. 2017;11:e0005968.
Wall KM, Kilembe W, Vwalika B, Dinh C, Livingston P, Lee Y-M, et al. Schistosomiasis is associated with incident HIV transmission and death in Zambia. PLoS Negl Trop Dis. 2018;12:e0006902.
Ssetaala A, Nakiyingi-Miiro J, Asiki G, Kyakuwa N, Mpendo J, Van Dam GJ, et al. Schistosoma mansoni and HIV acquisition in fishing communities of Lake Victoria, Uganda: a nested case-control study. Trop Med Int Health. 2015;20:1190–5.
Bochner AF, Baeten JM, Secor WE, Dam GJ, Szpiro AA, Njenga SM, et al. Associations between schistosomiasis and HIV‐1 acquisition risk in four prospective cohorts: a nested case‐control analysis. J Int AIDS Soc. 2020;23:e25534.
World Health Organization. Schistosomiasis. World Health Organization. 2020. https://www.who.int/news-room/fact-sheets/detail/schistosomiasis.
UNAIDS. The need for a holistic approach to women and HIV | UNAIDS. UNAIDS. 2020. https://www.unaids.org/en/resources/presscentre/featurestories/2018/march/20180316_holistic-approach-to-women-and-hiv.
Downs JA, de Dood CJ, Dee HE, McGeehan M, Khan H, Marenga A, et al. Schistosomiasis and human immunodeficiency virus in men in Tanzania. Am J Trop Med Hyg. 2017;96:856–62.
Dupnik K, Lee M, Mishra P, Reust M, Colombe S, Haider S, et al. Altered cervical mucosal gene expression and lower IL-15 levels in women with S. haematobium but not S. mansoni infection. J Infect Dis. 2019; 219:1777–85.
Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. 2017;46:29–37.
Anahtar M, Byrne E, Doherty K, Bowman B, Yamamoto H, Soumillon M, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42:965–76.
Masson L, Passmore J-AS, Liebenberg LJ, Werner L, Baxter C, Arnold KB. et al. Genital inflammation and the risk of HIV acquisition in women. Clin Infect Dis. 2015;61:260
Jenkins TP, Peachey LE, Ajami NJ, MacDonald AS, Hsieh MH, Brindley PJ, et al. Schistosoma mansoni infection is associated with quantitative and qualitative modifications of the mammalian intestinal microbiota. Sci Rep. 2018;8:12072.
Schneeberger PHH, Coulibaly JT, Panic G, Daubenberger C, Gueuning M, Frey JE, et al. Investigations on the interplays between Schistosoma mansoni, praziquantel and the gut microbiome. Parasit Vectors. 2018;11:168.
Kay GL, Millard A, Sergeant MJ, Midzi N, Gwisai R, Mduluza T, et al. Differences in the Faecal microbiome in schistosoma haematobium infected children vs. uninfected children. PLoS Negl Trop Dis. 2015;9:e0003861.
Ajibola O, Rowan AD, Ogedengbe CO, Mshelia MB, Cabral DJ, Eze AA, et al. Urogenital schistosomiasis is associated with signatures of microbiome dysbiosis in Nigerian adolescents. Sci Rep. 2019;9:829.
Adebayo AS, Suryavanshi MV, Bhute S, Agunloye AM, Isokpehi RD, Anumudu CI, et al. The microbiome in urogenital schistosomiasis and induced bladder pathologies. PLoS Negl Trop Dis. 2017;11:e0005826.
Gelfand M, Ross MD, Blair DM, Weber MC. Distribution and extent of schistosomiasis in female pelvic organs, with special reference to the genital tract, as determined at autopsy. Am J Trop Med Hyg. 1971;20:846–9.
Mishra P, Colombe S, Paul N, Mlingi J, Tosiri I, Aristide C, et al. Insufficiency of annual praziquantel treatment to control Schistosoma mansoni infections in adult women: a longitudinal cohort study in rural Tanzania. PLoS Negl Trop Dis. 2019;13:e0007844.
Berhe N, Medhin G, Erko B, Smith T, Gedamu S, Bereded D, et al. Variations in helminth faecal egg counts in Kato-Katz thick smears and their implications in assessing infection status with Schistosoma mansoni. Acta Trop. 2004;92:205–12.
Corstjens PLAM, van Lieshout L, Zuiderwijk M, Kornelis D, Tanke HJ, Deelder AM, et al. Up-converting phosphor technology-based lateral flow assay for detection of Schistosoma circulating anodic antigen in serum. J Clin Microbiol. 2008;46:171–6.
Corstjens PLAM, De Dood CJ, Kornelis D, Fat EMTK, Wilson RA, Kariuki TM, et al. Tools for diagnosis, monitoring and screening of Schistosoma infections utilizing lateral-flow based assays and upconverting phosphor labels. Parasitology. 2014;141:1841–55.
Ministry of Health and Social Welfare. The national road map strategic plan to accelerate reduction of maternal, newborn and child deaths in Tanzania (2016-2020). Dar es Salaam: Ministry of Health and Social Welfare; 2015.
Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E, Seok R, et al. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med. 2019;216:84–98.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4.
Edgar R. Local homology recognition and distance measures in linear time using compressed amino acid alphabets. Nucleic Acids Res. 2004;32:380–5.
Price JT, Vwalika B, Hobbs M, Nelson JAE, Stringer EM, Zou F, et al. Highly diverse anaerobe-predominant vaginal microbiota among HIV-infected pregnant women in Zambia. PLoS One. 2019;14:e0223128.
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59.
Navas-Molina JA, Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, et al. Advancing our understanding of the human microbiome using QIIME. Method Enzymol. 2013;531:371–444.
Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. Bioconductor Workflow for microbiome data analysis: from raw reads to community analyses. F1000Research. 2016;5:1492.
Kaul A, Mandal S, Davidov O, Peddada SD. Analysis of microbiome data in the presence of excess zeros. Front Microbiol. 2017;8:2114.
McClelland RS, Lingappa JR, Srinivasan S, Kinuthia J, John-Stewart GC, Jaoko W, et al. Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. Lancet Infect Dis. 2018;18:554–64.
Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. Nat Publ Group 2018;16:410–22.
Li H. Microbiome, metagenomics, and high-dimensional compositional data analysis. Annu Rev Stat Its Appl. 2015;2:73–94.
Polman K, De Vlas SJ, Gryseels B, Deelder AM. Relating serum circulating anodic antigens to faecal egg counts in Schistosoma mansoni infections: A modelling approach. Parasitology. 2000;121:601–10.
Borgdorff H, Tsivtsivadze E, Verhelst R, Marzorati M, Jurriaans S, Ndayisaba GF, et al. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in african women. ISME J. 2014;8:1781–93.
Nelson TM, Borgogna JLC, Brotman RM, Ravel J, Walk ST, Yeoman CJ. Vaginal biogenic amines: biomarkers of bacterial vaginosis or precursors to vaginal dysbiosis? Front Physiol. 2015;6:253.
Myer L, Kuhn L, Stein ZA, Wright TC, Denny L. Intravaginal practices, bacterial vaginosis, and women’s susceptibility to HIV infection: Epidemiological evidence and biological mechanisms. Lancet Infect Dis. 2005;5:786–94.
Lai SK, Hida K, Shukair S, Wang Y-Y, Figueiredo A, Cone R, et al. Human immunodeficiency virus type 1 is trapped by acidic but not by neutralized human Cervicovaginal Mucus. J Virol. 2009;83:11196–11200.
Taha TE, Hoover DR, Dallabetta GA, Kumwenda NI, Mtimavalye LAR, Yang LP, et al. Bacterial vaginosis and disturbances of vaginal flora: association with increased acquisition of HIV. AIDS. 1998;12:1699–706.
Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, et al. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis. 1999;180:1863–8.
Mikamo H, Kawazoe K, Izumi K, Ito K, Katoh N, Watanabe K, et al. Bacteriological epidemiology and treatment of bacterial vaginosis. Chemotherapy. 1996;42:78–84.
Agnew A, Fulford AJ, Mwanje MT, Gachuhi K, Gutsmann V, Krijger FW, et al. Age-dependent reduction of schistosome fecundity in Schistosoma haematobium but not Schistosoma mansoni infections in humans. Am J Trop Med Hyg. 1996;55:338–43.
Cheever AW, Kamel IA, Elwi AM, Mosimann JE, Danner R. Schistosoma mansoni and S. haematobium infections in Egypt. II. Quant parasitological Find necropsy Am J Trop Med Hyg. 1977;26:702–16.
Muller R. Worms and human disease. 2nd ed. CABI Publishing;UK 2001.
Turner JD, Narang P, Coles MCMA. Blood flukes exploit Peyer’s Patch lymphoid tissue to facilitate transmission from the mammalian host. Plos Pathog 2012;8:e1003063.
Wilson MS, Mentink-Kane MM, Pesce JT, Ramalingam TR, Thompson R, Wynn TA. Immunopathology of schistosomiasis. 2007;85:148–54.
Acknowledgements
We thank the study participants for their willing participation.
Funding
This work was supported by the Doris Duke Charitable Foundation [Grant number 2017067]; the Gilead Sciences Research Scholars Program in HIV from Gilead Sciences; and the National Institutes of Health [Grant number K23 AI 110238].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bullington, B.W., Lee, M.H., Mlingi, J. et al. Cervicovaginal bacterial communities in reproductive-aged Tanzanian women with Schistosoma mansoni, Schistosoma haematobium, or without schistosome infection. ISME J 15, 1539–1550 (2021). https://doi.org/10.1038/s41396-020-00868-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-00868-9
Keywords
This article is cited by
-
Metabolic profiles outperform the microbiota in assessing the response of vaginal microenvironments to the changed state of HPV infection
npj Biofilms and Microbiomes (2024)