Abstract
This study was conducted for the metagenomic analysis of stool samples from CRC affected individuals to identify biomarkers for CRC in Hainan, the only tropical island province of China. The gut microbiota of CRC patients differed significantly from that of healthy and reference database cohorts based on Aitchison distance and Bray–Cutis distance but there was no significant difference in alpha diversity. Furthermore, at the species level, 68 species were significantly altered including 37 CRC-enriched, such as, Fusobacterium nucleatum, Parvimonas micra, Gemella morbillorum, Citrobacter portucalensis, Alloprevotella sp., Shigella sonnei, Coriobacteriaceae bacterium, etc. Sixty-seven different metabolic pathways were acquired, and pathways involved in the synthesis of many amino acids were significantly declined. Besides, 2 identified antibiotic resistance genes performed well (area under the receive-operation curve AUC = 0.833, 95% CI 58.51–100%) compared with virulence factor genes. The results of the present study provide region-specific bacterial and functional biomarkers of gut microbiota for CRC patients in Hainan. Microbiota is considered as a non-invasive biomarker for the detection of CRC. Gut microbiota of different geographic regions should be further studied to expand the understanding of markers, especially for the China cohort due to diverse nationalities and lifestyles.




Similar content being viewed by others
Data Availability
The sequence data reported in this paper have been deposited in the NCBI database (metagenomic sequencing data: PRJNA608088, https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA608088). The analyses by R program can be found under: https://github.com/HNUmcc/CRC-cohort-Hainan-China. Further information and requests for resources and code should be directed to and will be fulfilled by the corresponding author (Jiachao Zhang, zhjch322123@163.com).
References
Torre LA et al (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108
Siegel RL, Jemal A, Ward EM (2009) Increase in incidence of colorectal cancer among young men and women in the United States. Cancer Epidemiol Biomarkers Prev 18(6):1695–1698
Levin B et al (2008) Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology 134(5):1570–1595
Turnbaugh PJ et al (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444(7122):1027–1031
Manichanh C et al (2006) Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55(2):205–211
Selgrad M et al (2008) The role of viral and bacterial pathogens in gastrointestinal cancer. J Cell Physiol 216(2):378–388
Louis P, Hold GL, Flint HJ (2014) The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12(10):661–672
Kostic AD et al (2013) Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14(2):207–215
Deschasaux M et al (2018) Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med 24(10):1526–1531
Murphy N et al (2019) Lifestyle and dietary environmental factors in colorectal cancer susceptibility. Mol Aspects Med 69:2–9
O’Keefe SJ (2016) Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol 13(12):691–706
Xie YH et al (2017) Fecal clostridium symbiosum for noninvasive detection of early and advanced colorectal cancer: test and validation studies. EBioMedicine 25:32–40
Butt J et al (2016) Association of Streptococcus gallolyticus subspecies gallolyticus with colorectal cancer: serological evidence. Int J Cancer 138(7):1670–1679
Mima K et al (2015) Fusobacterium nucleatum and T Cells in colorectal carcinoma. JAMA Oncol 1(5):653–661
Nosho K et al (2016) Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol 22(2):557–566
Boleij A et al (2015) The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 60(2):208–215
Pleguezuelos-Manzano C et al (2020) Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 580:269
Dai Z et al (2018) Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 6(1):70
Yang J et al (2020) Establishing high-accuracy biomarkers for colorectal cancer by comparing fecal microbiomes in patients with healthy families. Gut Microbes 11:1–12
Zhang J et al (2015) A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J 9(9):1979–1990
Stary L et al (2017) Are we any closer to screening for colorectal cancer using microbial markers? A critical review. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 161(4):333–338
Rosenbloom KR et al (2015) The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 43:670–681
Wood DE, Salzberg SL (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15(3):R46
Lu J et al (2017) Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3:104
Martín-Fernández J-A et al (2014) Bayesian-multiplicative treatment of count zeros in compositional data sets. Stat Model. https://doi.org/10.1177/1471082X14535524
Franzosa EA et al (2018) Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15(11):962–968
Jia B et al (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(D1):D566–D573
Liu B et al (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47(D1):D687–D692
Langmead B et al (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25
Zapala MA, Schork NJ (2006) Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc Natl Acad Sci USA 103(51):19430–19435
Wirbel J et al (2019) Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 25(4):679–689
Liang Q et al (2017) Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res 23(8):2061–2070
Mizutani S, Yamada T, Yachida S (2020) Significance of the gut microbiome in multistep colorectal carcinogenesis. Cancer Sci 111:766
Song M, Chan AT, Sun J (2020) Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology 158(2):322–340
Kwong TNY et al (2018) Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology 155(2):383–390
Igbinosa EO et al (2018) Draft genome sequence of multidrug-resistant strain Citrobacter portucalensis MBTC-1222, isolated from Uziza (Piper guineense) leaves in Nigeria. Genome Announc 6:9
Hasan MS, Sultana M, Hossain MA (2019) Complete genome arrangement revealed the emergence of a poultry origin superbug Citrobacter portucalensis strain NR-12. J Glob Antimicrob Resist 18:126–129
Tilg H et al (2018) The intestinal microbiota in colorectal cancer. Cancer Cell 33(6):954–964
Queen J, Zhang J, Sears CL (2020) Oral antibiotic use and chronic disease: long-term health impact beyond antimicrobial resistance and Clostridioides difficile. Gut Microbes 11:1–12
Duan Y et al (2020) Gut resistomes, microbiota and antibiotic residues in Chinese patients undergoing antibiotic administration and healthy individuals. Sci Total Environ 705:135674
Kashani N et al (2020) FadA-positive Fusobacterium nucleatum is prevalent in biopsy specimens of Iranian patients with colorectal cancer. New Microbes New Infect 34:100651
Acknowledgements
We sincerely thank all the volunteers for their participation.
Funding
This work was supported by Key R & D programs in Hainan (Grant No. ZDYF2019150), the Scientific Research Foundation of Hainan University (Grant No. KYQD1548).
Author information
Authors and Affiliations
Contributions
The study was designed by JZ and KNC. The experiment was performed by CM, KC. Data collection was performed by HC, YT, YW and QO. Data analysis was performed by CM, RM and KC. The manuscript was written by CM, RM, KC, SW, JZ and KNC. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no competing interests.
Ethical Approval
The protocol for the study was approved by the Ethics Committee Hainan University.
Informed Consent
All the participants in the study were informed about the study and were provided with written consent. Sampling and all described subsequent steps were conducted in accordance with the approved guidelines.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file1 (TIF 2004 KB)
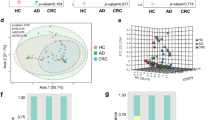
AUC based on the markers of VFGs classified HN_CRC and Con group
Rights and permissions
About this article
Cite this article
Chang, H., Mishra, R., Cen, C. et al. Metagenomic Analyses Expand Bacterial and Functional Profiling Biomarkers for Colorectal Cancer in a Hainan Cohort, China. Curr Microbiol 78, 705–712 (2021). https://doi.org/10.1007/s00284-020-02299-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02299-3