Abstract
Introduction
Human rhinovirus (HRV) is one of the most common human viral pathogens related to infections of the upper and lower respiratory tract, which can result in bronchiolitis and pneumonia. However, the relevance of HRV in human health was under-estimated for long time due to the absence of molecular targets for influenza and influenza-like syndrome surveillance in Brasília, Brazil.
Objectives
The main objective of this study was analyze the clinical characteristics and outcomes of HRV infections in comparison with patients without HRV and other common respiratory viruses.
Materials and Methods
For this purpose, new specific primer sets were designed based on the high throughput sequencing analysis in previous study. These primers were used for HRV detection by RT-qPCR and Sanger sequencing of amplified cDNA of 5′ genomic region. The phylogenetic tree with representative HRV isolates was constructed using the Mega X software. Statistical analysis considering the patient profiles were performed using IBM SPSS program with non-parametric tests.
Results
The most prevalent virus in negative samples was rhinovirus (n = 40), including three rhinovirus species (rhinovirus A, B, and C). The odds ratio associated with HRV infection was 2.160 for patients younger than 2 years and 4.367 for people living in rural areas. The multiple analysis showed lower chance of patients with HRV presenting respiratory distress.
Conclusion
In this study, it was reported the predominance of rhinoviruses in cases of respiratory illness for negative patients for the influenza and influenza-like syndrome surveillance, being rhinorrhea, the most significant symptom associated with the disease.
Similar content being viewed by others
Introduction
Human rhinovirus (HRV) is one of the most common human viral pathogens. They are non-enveloped single-stranded RNA virus, member of Enterovirus genus, of Picornaviridae family [1, 2]. About half of common cold are caused by rhinoviruses, which have more than 160 serotypes described, in three species (rhinovirus A, B, and C) [3]. In addition, cold leads to economic problems related clinical morbidity, overloading the health system, also causing absences at work and in schools [2, 4].
Rhinoviruses cause not only upper respiratory tract infections but also lower respiratory tract infections, mainly in children and patients with chronic pulmonary disease or immunocompromised [5]. The most common symptoms of upper infection are sore throat, rhinorrhea, headache, cough, malaise, and lower fever. However, in some cases, there may be aggravation for rhinosinusitis and otitis media [6, 7]. Infections of the lower respiratory tract are often associated with bronchiolitis and pneumonia especially in children [8,9,10].
In Brazil, the implementation of the sentinel surveillance system began in 2000 [11], so referral hospitals have been conducting active surveillance to detect respiratory viruses. Such surveillance includes notification and laboratory investigation of cases with the diagnostic criteria of influenza-like syndrome (ILS) and severe acute respiratory syndrome (SARS). This viral respiratory infection monitoring has resulted in important information about the circulation of community-acquired respiratory viruses. However, some viruses may not be identified by the absence of RT-qPCR target performed at central laboratories.
The present study reports the molecular and clinical characteristics, and outcomes of HRV infections in comparison with patients without HRV and other common respiratory viruses.
Materials and methods
Detection of HRV by RT-qPCR
The nasopharyngeal and tracheal secretion samples received in the laboratory in June, July, and August 2016 were tested for the respiratory virus diagnostic panel performed in Lacen-DF (Central Laboratory at Federal District, Brasilia, Brazil) by the standard RT-qPCR [12] (for influenza A virus, influenza B virus, human orthopneumovirus, human metapneumovirus, human mastadenovirus C, human respirovirus 1, human orthorubulavirus 2, and human respirovirus 3). The target genes of the standard RT-qPCR are shown in the Supplementary Table 1. During transport, the samples were stored on ice and sent to the laboratory within 48 h after collection. Those that had negative results were separated and stored in freezer at − 70 °C for subsequent DNA/RNA extraction. The metagenomic analyses by high throughput sequencing (HTS) of those samples as one pooled virome were previously studied [13]. Based on this study, HRV universal primers targeting conserved regions in 5’ UTR (untranslated region) were newly designed for HRV detection (Table 1) using the Geneious R8.1 program (Biomatters, Auckland, New Zealand). The RNA was extracted from 145 samples using the Magna Pure LC Total Nucleic Acid Kit (Hoffmann-La Roche, Basel, Switzerland), and then all the 145 samples were projected to the RT-qPCR using GoTaq® Probe 1-Step RT-qPCR System (Promega, Madison, USA). Positivity for rhinovirus was defined with cycle threshold (CT) less than or equal to 40 (total of 45 cycles in the reaction) [12]. Human RNase P gene was used for endogenous control for the same sample set in separate tube.
RT-PCR and sanger sequencing
The HRV_Com_For and HRV_Com_Rev primers were newly designed (Table 1) targeting conserved regions based on virome analyses by HTS. This primer pair was used to amplify an approximately 540-bp fragment for amplicon sequencing, including part of 5’ UTR and the VP4/VP2 protein gene of HRV A, B, and C. cDNA of the selected samples were synthesized using MMLV transcriptase (Thermo Fisher Scientific, Waltham, USA) with random hexamer primer and, then, amplified with LongAmp Taq DNA Polymerase (New England BioLabs, Ipswich, USA). The PCR product was purified and sequenced by the Sanger method at Macrogen Inc. (Seoul, South Korea).
Sequence analysis
The nucleotide (nt) sequences were determined using the Geneious R8.1 program (Biomatters, Auckland, New Zealand). Complete rhinovirus genome sequences were retrieved from GenBank as references and aligned with the sequences obtained in this study. This alignment was again trimmed to set the sequence size determined in this study (approximately 500 bases, the size has small variation due to indel mutation). The phylogenetic tree based on the 5’ UTR and the VP4/VP2 protein gene was constructed using the Mega X software [14]. The tree was composed by eight nucleotide sequences from this study and other forty rhinovirus isolates, that were complete genome sequences available on databases. Phylogenetic tree was inferred using maximum likelihood method with Hasegawa-Kishino-Yano model as indicated by jModelTest analysis [15]. In order to obtain more precise nt identity in pairwise comparison, three more related virus sequences for each sequence in this study were retrieved by BlastN search.The pairwise comparison was then performed.
Statistical analysis
Medical records were obtained from compulsory notification sheet for ILS and SARS sent with the sample to the laboratory. Data were analyzed using the software IBM SPSS (Chicago, USA). Non-parametric tests were used as appropriate, for qualitative and quantitative variables. All p values were two tailed and a value of < 0.05 was considered significant.
Results
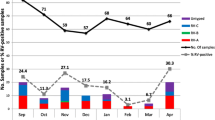
Rhinoviruses were identified as the main virus found in negative samples for the regular respiratory virus survey in Lacen-DF, including influenza A virus, influenza B virus, human orthopneumovirus, human metapneumovirus, human mastadenovirus C, human respirovirus 1, human orthorubulavirus 2, and human respirovirus 3. The RT-qPCR using newly designed primers and probe (Table 1) detected HRV in 40 out of 145 samples analyzed. The mean CT scan was about 28 ranging from 17 to 39. These primers targeted the 5’ UTR region (Table 1), which is most conserved among rhinoviruses.
To identify HRV at species level, primers which target a region of approximately 540 bp of the 5’ UTR-VP4/VP2 protein gene region were sequenced. Five samples were concluded of HRV-A, one of HRV-B and two of HRV-C (Fig. 1), out of eight sequenced samples of the patients in the Federal District during the study period. The other samples were not of sufficient quality for sequencing. All sequences were deposited in the databank: rhinovirus A BSB_8 (Accession number LC549198), rhinovirus A BSB_16 (LC549199), rhinovirus A BSB_26 (LC549200), rhinovirus A BSB_33 (LC549201), rhinovirus A BSB_38 (LC549202), rhinovirus B BSB_30 (LC549203), rhinovirus C BSB_2 (LC549204), and rhinovirus C BSB_15 (LC549205). It is interesting to note that although a small number of samples were sequenced, the genetic variability of HRV-A was very wide, positioning in five different clusters in the phylogenetic tree (Fig. 1). Among them, only nt identities of 72.94–83.86% were observed (Table 2). On the other hand, two isolates of HRV-C were very similar, possessing 98.99% of nt identity.
Phylogenetic tree of rhinoviruses based on 5’ UTR and the VP4 / VP2 protein gene (approximately 500 bases). The tree was constructed using Hasegawa-Kishino-Yano model of the maximum likelihood method with 48 isolates, which eight were from the Federal District, Brazil, and the other 40 were genome sequences available on databases. Bootstrap values above 50 (500 repetitions), and accession number of each sequence are indicated. Bar: number of substitutions per site. Green circle: Rhinovirus A, blue circle: Rhinovirus B, and red circle: Rhinovirus C. The identification of isolate from the Federal District, Brazil are highlighted with colors used for species indication
When identifying the location of the most related virus (in the databases), it can be observed that there is almost no relation in the geographical distribution (Table 3). The most related rhinovirus isolates came from several continents such as America, Asia, Europe, and Oceania. This observation suggests that the dissemination of the rhinoviruses can happen worldwide in long distance and is different from the arbovirus geographical dissemination.
Patients with (n = 40) and without (n = 105) rhinovirus were compared in relation to socio-demographic and clinical parameters in order to evaluate possible risk factors associated with the involvement of these viruses. It can be observed in Table 4 that patients with rhinovirus were significantly different from patients negative for the main respiratory viruses in relation to age (P = 0.042) and to the area of residence (P = 0.019). Patients younger than 2 years presented 2.160 times more likely to be infected by rhinovirus than patients with 2 years old or more.
In this study, patients with rhinovirus were significantly younger than patients without rhinovirus (P = 0.042) (Table 5). The median age for patients with the virus was 2.50 (interquartile range = 27.38) versus 26.00 (interquartile range = 50.13) years for patients without this virus. Thus, greater susceptibility of children to rhinovirus infection is reported. Figure 2 shows the difference between the groups with and without the virus in relation to the days of symptoms and age. It is observed that the days of symptoms did not differ significantly, but the age distribution of the patients was significantly lower in patients who presented rhinovirus respiratory symptoms (P = 0.042).
Box-plot of days of symptoms (a) and age (b) of patients with acute respiratory infection treated in the Federal District, Brazil, 2016. The comparison shows patients with and without rhinovirus infection, where only the age difference was statistically significant (P = 0.042). Extreme outliers are marked with an asterisk and moderate outliers with a dot
Patients with rhinovirus were located significantly more in rural areas when compared to negative patients. The odds ratio for the rural area was 4.367 (1/0.229), which means that people resident in rural areas were 4.367 times more affected by rhinovirus when compared to urban dwellers.
Regarding the main symptoms of patients with acute respiratory infection, it can be observed in Table 6 that patients with rhinovirus presented significantly more rhinorrhea in relation to the patients without this virus detected (P = 0.001). Patients with rhinovirus were 3.981 times more likely to presenting rhinorrhea in relation to negative patients. There was no statistically significant difference in relation to the other symptoms.
A multiple logistic regression with 20 explanatory variables (age, days of symptoms, month, diagnostic criteria, sex, breed, zone, risk factor, vaccinated, hospitalization, medication, use of ventilatory support, fever, cough, sore throat, dyspnea, respiratory distress, myalgia, O2 saturation < 95%, and rhinorrhea) was performed to analyze the variables associated with rhinovirus infection (response variable). In the model containing only the constant, there was 71% agreement with the classification table. When entering the explanatory variables, the concordance increased to 79.8%.
The regression model was statistically significant (P = 0.005), with adequate adjustments (P = 0.862—Hosmer’s test and Lemeshow) and R2 = 0.391. To improve the model, a technique of variable selection was used by the backwise stepwise method. The best fit was obtained after 15 steps of exclusion of variables, remaining six explanatory variables. The new model was also statistically significant (P < 0.001), with optimal adjustment (P = 0.923—Hosmer and Lemeshow test) and R2 = 0.319. Three significant explanatory variables were obtained: respiratory distress, rhinorrhea, and medication (Table 7).
Patients with rhinovirus presented 2.985 (1/0.335) times less chance of experiencing respiratory distress, 6.601 and 3.892 times more chance of having rhinorrhea and using medication (oseltamivir), respectively. Thus, it is observed that the main symptoms related to rhinovirus infection were the presence of rhinorrhea and absence of respiratory discomfort. The use of antiviral medication, oseltamivir, was also significantly more observed in patients with rhinovirus in the multiple analysis.
To assess how respiratory symptoms were associated in patients with rhinovirus infection, a cluster analysis was performed with the 40 positive patients, using dendrogram with Ward method and measure of the quadratic Euclidean distance. It is observed that two groups of symptoms were mainly formed: fever, cough, and rhinorrhea in the first group and myalgia, O2 saturation < 95%, respiratory distress, dyspnea, and sore throat in the second group (Fig. 3). The symptoms of the first group may be associated with cases of common cold (upper airways infection), and those of the second group with cases of complications of infection in the lower respiratory tract.
To investigate whether age was not a covariate of rhinorrhea, the Mantel-Haenszel test was performed, which evaluated the association between rhinorrhea symptom and age-corrected rhinovirus. It was observed that the association between rhinorrhea and rhinovirus occurred in the group of patients older than 2 years (P = 0.001), but not in the group younger than 2 years (P = 1,000) (Supplementary Table 2); that is, children with and without rhinovirus presented rhinorrhea in a similar way. The Mantel-Haenszel test showed odds ratio = 3.865 (1.483–10.076; P = 0.005), which shows that even if adjusted for age, rhinorrhea was statistically significant to the rhinovirus, where rhinovirus patients had 3.865 times more chance of presenting this symptom compared with patients without the virus.
Discussion
Since the beginning of the monitoring of respiratory virus circulation in the country, rhinovirus has been found with high frequency, either alone or co-detected with other respiratory viruses, mainly in the south and southeast regions [16, 17]. To date, however, the circulation of these viruses in the central region of the country has not been well studied. Therefore, the identification of these viruses is crucial to determine the etiologic agent of respiratory illness. In this case, the 5’ UTR region is the most suitable to identify the virus by RT-qPCR, and this region has already been used by other laboratories with similar oligonucleotides [18, 19].
Rhinoviruses are reported to be the main cause of the common cold and spread from person to person via direct and indirect contact, mainly by infected respiratory secretions. The transmission by hands is important in this context [20]. The easy contagion of rhinovirus can explain its rapid spread, over long distances, as shown in Table 3.
HRV infections were associated with diseases such as asthma, chronic pulmonary disease, bronchiolitis, and pneumonia in children, the elderly, and immunocompromised. Thus, proper clinical management, timely diagnosis, and containment of outbreaks are becoming important [5, 16].
Rhinovirus infection was significantly associated with residency in rural areas. In this study, 7% came from rural areas, and among those infected by the virus, the percentage was 15%. A study in Brazil has already reported that children from urban areas of school age are more susceptible to respiratory inflammatory problems compared to those in rural regions [21]. Thus, the negative cases for rhinovirus and other common respiratory viruses may be associated with allergic inflammatory processes, which is more frequent in urban areas. With this, rural patients would present respiratory problems related mainly to viral infections, as observed in relation to HRV. In the multiple analysis, the relationship between rural residence and rhinovirus was not observed.
Patients younger than 2 years were more subject to HRV infection and had a significantly lower proportion of cases with fever, and more comorbidities such as asthma than patients without HRV [16, 22, 23]. In this study, it was observed that children younger than 2 years were more susceptible to rhinovirus infection, but there was no association with comorbidities. The main symptom associated with HRV was rhinorrhea rather than low frequency of fever. The HRV infections predominated during autumn and winter with significant negative correlation between the number of HRV cases and the average temperature [16]. So, the result obtained in this study, with rhinovirus dominance in negative samples for the other common respiratory viruses, was expected since the samples were collected in late fall and winter (June, July, and August), when the virus circulation is high.
The data here presented showed that patients with rhinovirus were significantly younger than patients without rhinovirus, consistent with other works that showed the detection rate of HRV infection in children was significantly higher than that in adults, mainly the HRV-C. Clinical manifestations associated with HRV-C appear to be more severe in children [24] and often associated with respiratory complications, like acute wheezing illness and bronchiolitis [25, 26]. In adults, HRV-A is more common and is associated with cases of viral pneumonia [27].
Fever and nasal congestion were more frequently observed in pediatric group than the adults which were most diagnosed with severe respiratory disease entity [28]. Figure 3 presented two groups of symptoms. This division may be associated with the main affected area, upper or lower respiratory tract. Rhinorrhea was the clinical manifestation significantly associated with HRV infection in this study. Other studies also show rhinorrhea as the main symptom associated with rhinovirus infection [29, 30].
The use of oseltamivir was significantly associated with HRV infection in the multiple analysis (Table 7). In univariate analysis, this association was not verified (Table 4). However, a high percentage (37.5%) of patients with rhinoviruses who took the medication incorrectly were observed. Although rhinovirus and influenza symptoms can be similar and therefore confounded [31], the use of oseltamivir in patients with HRV has already been reported, but in a much smaller percentage (2.1–2.5%) [22, 31].
Conclusion
This study showed the predominance of rhinoviruses in cases of respiratory illness for negative patients for the influenza and influenza-like syndrome surveillance. Rhinorrhea was the most significant symptom associated with the disease. All three virus species were identified in the Federal District, Brazil. A lower chance of presenting respiratory distress was also observed in the multiple analysis, although the symptoms of patients with HRV were associated with both upper airways infection and complications of infection in the lower respiratory tract (Fig. 3). In the patients with HRV, 55.0 and 37.5% presented exacerbations such as dyspnea and respiratory distress, and 57.5% had to be hospitalized (Tables 4 and 6). As HRV is also identified in asymptomatic patients [32, 33], more detailed studies need to be made to better understand the prevalence, risk factors, and outcomes related to rhinovirus infection in the population.
Availability of data and material
The data for this study were made available by the Health Department of the Federal District through a project partnership and approved by an ethics committee. All genetic sequences of rhinovirus were deposited in the databank.
References
Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser-Liggett CM, Liggett SB (2009) Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 324:55–59
Gern JE (2010) The ABCs of rhinoviruses, wheezing, and asthma. J Virol 84:7418–7426
Palmenberg AC, Rathe JA, Liggett SB (2010) Analysis of the complete genome sequences of human rhinovirus. J Allergy Clin Immunol 125:1190–1201
Fendrick AM, Monto AS, Nightengale B, Sarnes M (2003) The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch Intern Med 163:487–494
Jacobs SE, Lamson DM, George KS, Walsh TJ (2013) Human rhinoviruses. Clin Microbiol Rev 26:135–162
Blomqvist S, Roivainen M, Puhakka T, Kleemola M, Hovi T (2002) Virological and serological analysis of rhinovirus infections during the first two years of life in a cohort of children. J Med Virol 66:263–268
Turner BW, Cail WS, Hendley JO et al (1992) Physiologic abnormalities in the paranasal sinuses during experimental rhinovirus colds. J Allergy Clin Immunol 90:474–478
Henquell C, Mirand A, Deusebis A-L, Regagnon C, Archimbaud C, Chambon M, Bailly JL, Gourdon F, Hermet E, Dauphin JB, Labbé A, Peigue-Lafeuille H (2012) Prospective genotyping of human rhinoviruses in children and adults during the winter of 2009-2010. J Clin Virol 53:280–284
Lieberman D, Shimoni A, Shemer-Avni Y, Keren-Naos A, Shtainberg R, Lieberman D (2010) Respiratory viruses in adults with community-acquired pneumonia. Chest 138:811–816
Broberg E, Niemelä J, Lahti E, Hyypiä T, Ruuskanen O, Waris M (2011) Human rhinovirus C-associated severe pneumonia in a neonate. J Clin Virol 51:79–82
Barros FR Daufenbach LZ, Vicente MG et al (2004) O desafio da influenza: epidemiologia e organização da vigilância no Brasil. Boletim Eletrônico Epidemiológico da Secretaria de Vigilância em Saúde, [S.l.], v. 1, p. 1–7. Available from: http://portalsaude.saude.gov.br/images/pdf/2014/julho/16/Ano04-n01-desafioinfluenza-br-completo.pdf. Accessed 13 July 2020
World Health Organization. (2009) CDC protocol of realtime RTPCR for influenza A (H1N1). Available from: http://www.who.int/csr/resources/publications/swineflu/realtimeptpcr/en/index.html. Accessed 25 July 2020
Souza LC, Blawid R, Silva JMF, Nagata T (2019) Human virome in nasopharynx and tracheal secretion samples. Mem Inst Oswaldo Cruz 114:e190198
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Leotte J, Trombetta H, Faggion HZ, Almeida BM, Nogueira MB, Vidal LR, Raboni SM (2017) Impact and seasonality of human rhinovirus infection in hospitalized patients for two consecutive years. J Pediatr 93:294–300
Watanabe ASA, Carraro E, Candeias JMG, Donalísio MR, Leal É, Granato CFH, Bellei N (2011) Viral etiology among the elderly presenting acute respiratory infection during the influenza season. Rev Soc Bras Med Trop 44:18–21
Reijans M, Dingemans G, Klaassen CH, Meis JF, Keijdener J, Mulders B, Eadie K, van Leeuwen W, van Belkum A, Horrevorts AM, Simons G (2008) RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol 46:1232–1240
Jaramillo-Gutierrez G, Benschop KS, Claas EC et al (2013) September through October 2010 multi-centre study in the Netherlands examining laboratory ability to detect enterovirus 68, an emerging respiratory pathogen. J Virol Methods 190:53–62
Gwaltney JM Jr, Moskalski PB, Hendley JO (1978) Hand-to-hand transmission of rhinovirus colds. Ann Intern Med 88:463–467
Galvão CE, Saldiva PH, Kalil Filho JE, Castro FF (2004) Inflammatory mediators in nasal lavage among school-age children from urban and rural areas in São Paulo, Brazil. São Paulo Med J 122:204–207
Tam PI, Zhang L, Cohen Z (2018) Clinical characteristics and outcomes of human rhinovirus positivity in hospitalized children. Ann Thorac Med 13:230–236
Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, Martin AC, Khoo SK, Cox DW, Geelhoed GC, McMinn PC, Goldblatt J, Gern JE, le Souef PN (2011) Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J 37:1037–1042
Lee WM, Lemanske RF Jr, Evans MD et al (2012) Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med 186:886–891
Tapparel C, Junier T, Gerlach D, van Belle S, Turin L, Cordey S, Mühlemann K, Regamey N, Aubert JD, Soccal PM, Eigenmann P, Zdobnov E, Kaiser L (2009) New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis 15:719–726
Wisdom A, Kutkowska AE, McWilliam Leitch EC et al (2009) Genetics, recombination and clinical features of human rhinovirus species C (HRV-C) infections; interactions of HRV-C with other respiratory viruses. PLoS One 4:e8518
Linder JE, Kraft DC, Mohamed Y, Lu Z, Heil L, Tollefson S, Saville BR, Wright PF, Williams JV, Miller EK (2013) Human rhinovirus C: age, season, and lower respiratory illness over the past 3 decades. J Allergy Clin Immunol 131:69–77
Hung HM, Yang SL, Chen CJ, Chiu CH, Kuo CY, Huang KYA, Lin TY, Hsieh YC, Gong YN, Tsao KC, Huang YC (2019) Molecular epidemiology and clinical features of rhinovirus infections among hospitalized patients in a medical center in Taiwan. J Microbiol Immunol Infect 52:233–241
Lethbridge R, Prastanti F, Robertson C, Oo S, Khoo SK, le Souëf PN, Laing IA (2018) Prospective assessment of rhinovirus symptoms and species recurrence in children with and without an acute wheezing exacerbation. Viral Immunol 31:299–305
Annamalay AA, Khoo SK, Jacoby P, Bizzintino J, Zhang G, Chidlow G, Lee WM, Moore HC, Harnett GB, Smith DW, Gern JE, LeSouef P, Laing IA, Lehmann D, Kalgoorlie Otitis Media Research Project Team (2012) Prevalence of and risk factors for human rhinovirus infection in healthy aboriginal and non-aboriginal Western Australian children. Pediatr Infect Dis J 31:673–679
Boivin G, Osterhaus AD, Gaudreau A, Jackson HC, Groen J, Ward P (2002) Role of picornaviruses in flu-like illnesses of adults enrolled in an oseltamivir treatment study who had no evidence of influenza virus infection. J Clin Microbiol 40:330–334
Iwane MK, Prill MM, Lu X, Miller EK, Edwards KM, Hall CB, Griffin MR, Staat MA, Anderson LJ, Williams JV, Weinberg GA, Ali A, Szilagyi PG, Zhu Y, Erdman DD (2011) Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J Infect Dis 204:1702–1710
Byington CL, Ampofo K, Stockmann C, Adler FR, Herbener A, Miller T, Sheng X, Blaschke AJ, Crisp R, Pavia AT (2015) Community surveillance of respiratory viruses among families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) Study. Clin Infect Dis 61:1217–1224
Funding
This work was supported by grant from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) with the project number of 309670/2018-7.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by all the authors. The first draft of the manuscript was written by Larissa da Costa Souza and Tatsuya Nagata and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
This research project was reviewed and approved by ethics committee of the Faculty of Health Sciences (University of Brasília), approval letter No. 3.052.443.
Consent to participate
This study obtained waiver of the informed consent form.
Consent for publication
Not applicable
Code availability
Not applicable.
Additional information
Responsible Editor: Giliane Souza Trindade
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
da Costa Souza, L., Bello, E.J.M., dos Santos, E.M. et al. Molecular and clinical characteristics related to rhinovirus infection in Brasília, Brazil. Braz J Microbiol 52, 289–298 (2021). https://doi.org/10.1007/s42770-020-00411-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-020-00411-0