Abstract
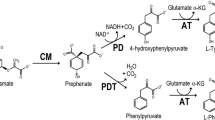
A putative aromatic amino acid ammonia-lyase gene (named Pl-pal) was discovered in Photorhabdus luminescens DSM 3368. BLAST and phylogenetic analyses predicted that this enzyme is a histidine ammonia-lyase, whereas sequence alignment suggested that it is more likely a phenylalanine ammonia-lyase (PAL). This gene was amplified from P. luminescens and expressed in Escherichia coli BL21(DE3). The function of Pl-PAL (58 kDa) was characterized by in vitro enzymatic reactions with l-phenylalanine (l-Phe), l-tyrosine (l-Tyr), l-histidine (l-His), and l-tryptophan (l-Trp). Pl-PAL can convert l-Phe and l-Tyr to trans-cinnamic acid and p-coumaric acid, respectively, but had no function on l-His and l-Trp. The optimum temperature and pH were determined to be 40 °C and 11.0, respectively. Under the optimal conditions, Pl-PAL had a kcat/Km value of 0.52 s−1 mM−1 with l-Phe as the substrate, while only 0.013 s−1 mM−1 for l-Tyr. Therefore, the primary function of Pl-PAL was determined to be PAL. The Pl-pal-harboring E. coli strain was used as a whole-cell biocatalyst to produce trans-cinnamic acid from l-Phe. The overall molar conversion rate and productivity were 65.98% and 228.10 mg L−1 h−1, respectively, after the cells were repeatedly utilized 7 times. This work thus provides a promising strain for industrial production of trans-cinnamic acid.








Similar content being viewed by others
Data availability
All relevant data are included in this paper. The nucleotide and protein sequences can be accessed in NCBI GenBank based on the accession numbers. Materials are available upon reasonable request to the corresponding author.
References
Otto, M., Wynands, B., Lenzen, C., Filbig, M., Blank, L. M., & Wierckx, N. (2019). Rational engineering of phenylalanine accumulation in Pseudomonas taiwanensis to enable high-yield production of trans-cinnamate. Frontiers in Bioengineering and Biotechnology, 7, 312. https://doi.org/10.3389/fbioe.2019.00312.
van Summeren-Wesenhagen, P. V., & Marienhagen, J. (2015). Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin. Applied and Environmental Microbiology, 81(3), 840–849. https://doi.org/10.1128/AEM.02966-14.
Gunia-Krzyżak, A., Słoczyńska, K., Popiół, J., Koczurkiewicz, P., Marona, H., & Pękala, E. (2018). Cinnamic acid derivatives in cosmetics current use and future prospects. International Journal of Cosmetic Science, 40(4), 356–366. https://doi.org/10.1111/ics.12471.
Sirichai, A. (2017). Cinnamic acid and its derivatives: mechanisms for prevention and management of diabetes and its complications. Nutrients, 9(2), 163. https://doi.org/10.3390/nu9020163.
Flourat, A. L., Combes, J., Bailly-Maitre-Grand, C., Magnien, K., Haudrechy, A., Renault, J.-H., & Allais, F. Accessing p-hydroxycinnamic acids: chemical synthesis, biomass recovery, or engineered microbial production? ChemSusChem. https://doi.org/10.1002/cssc.202002141.
Sharma, P. (2011). Cinnamic acid derivatives: a new chapter of various pharmacological activities. Journal of Chemical and Pharmaceutical Research, 3(2), 403–423.
Weise, N. J., Parmeggiani, F., Ahmed, S. T., & Turner, N. J. (2018). Discovery and investigation of mutase-like activity in a phenylalanine ammonia lyase from Anabaena variabilis. Topics in Catalysis, 61(3), 288–295. https://doi.org/10.1007/s11244-018-0898-1.
Lin, W., Liu, A., Weng, C., Li, H., Sun, S., Song, A., & Zhu, H. (2017). Cloning and characterization of a novel phenylalanine ammonia-lyase gene from Inonotus baumii. Enzyme and Microbial Technology, 112, 52–58. https://doi.org/10.1016/j.enzmictec.2017.10.010.
Nagy, E. Z. A., Tork, S. D., Lang, P. A., Filip, A., & Bencze, L. C. (2019). Mapping the hydrophobic substrate binding site of phenylalanine ammonia lyase from Petroselinum crispum. ACS Catalysis, 9(9), 8825–8834. https://doi.org/10.1021/acscatal.9b02108.
Xu, F., Cai, R., Cheng, S., Du, H., & Cheng, S. (2008). Molecular cloning, characterization and expression of phenylalanine ammonia-lyase gene from Ginkgo biloba. African Journal of Biotechnology, 7(6), 721–729.
Tork, S. D., Nagy, E. Z. A., Cserepes, L., Bordea, D. M., Nagy, B., Toşa, M. I., Paizs, C., & Bencze, L. C. (2019). The production of L- and D-phenylalanines using engineered phenylalanine ammonia lyases from Petroselinum crispum. Scientific Reports, 9(1), 20123. https://doi.org/10.1038/s41598-019-56554-0.
Hardegger, L. A., Beney, P., Bixel, D., Fleury, C., Gao, F., Perrenoud, A. G.-G., Gu, X., Haber, J., Hong, T., Humair, R., Kaegi, A., Kibiger, M., Kleinbeck, F., Luu, V. T., Padeste, L., Rampf, F. A., Ruch, T., Schlama, T., Sidler, E., Udvarhelyi, A., Wietfeld, B., & Yang, Y. (2020). Toward a scalable synthesis and process for EMA401, Part III: Using an engineered phenylalanine ammonia lyase enzyme to synthesize a non-natural phenylalanine derivative. Organic Process Research & Development, 24(9), 1763–1771. https://doi.org/10.1021/acs.oprd.0c00217.
Vargas-Tah, A., Martínez, L., Hernández-Chávez, G., Rocha, M., Martínez, A., Bolívar, F., & Gosset, G. (2015). Production of cinnamic and p-hydroxycinnamic acid from sugar mixtures with engineered Escherichia coli. Microbial Cell Factories, 14(1), 6. https://doi.org/10.1186/s12934-014-0185-1.
Bang, H. B., Lee, K., Lee, Y. J., & Jeong, K. J. (2018). High-level production of trans-cinnamic acid by fed-batch cultivation of Escherichia coli. Process Biochemistry, 68, 30–36. https://doi.org/10.1016/j.procbio.2018.01.026.
Noda, S., Miyazaki, T., Miyoshi, T., Miyake, M., Okai, N., Tanaka, T., Ogino, C., & Kondo, A. (2011). Cinnamic acid production using Streptomyces lividans expressing phenylalanine ammonia lyase. Journal of Industrial Microbiology & Biotechnology, 38(5), 643–648. https://doi.org/10.1007/s10295-011-0955-2.
Appert, C., Logemann, E., Hahlbrock, K., Schmid, J., & Amrhein, N. (1994). Structural and catalytic properties of the four phenylalanine ammonia-lyase isoenzymes from parsley (Petroselinum crispum Nym.). European Journal of Biochemistry, 225(1), 491–499. https://doi.org/10.1111/j.1432-1033.1994.00491.x.
Wang, S., Zhang, S., Zhou, T., Zeng, J., & Zhan, J. (2013). Design and application of an in vivo reporter assay for phenylalanine ammonia-lyase. Applied Microbiology and Biotechnology, 97(17), 7877–7885. https://doi.org/10.1007/s00253-013-5122-4.
Dreßen, A., Hilberath, T., Mackfeld, U., Billmeier, A., Rudat, J., & Pohl, M. (2017). Phenylalanine ammonia lyase from Arabidopsis thaliana (AtPAL2): a potent MIO-enzyme for the synthesis of non-canonical aromatic alpha-amino acids: Part I: Comparative characterization to the enzymes from Petroselinum crispum (PcPAL1) and Rhodosporidium toruloides (RtPAL). Journal of Biotechnology, 258, 148–157. https://doi.org/10.1016/j.jbiotec.2017.04.005.
Röther, D., Poppe, L., Viergutz, S., Langer, B., & Rétey, J. (2001). Characterization of the active site of histidine ammonia-lyase from Pseudomonas putida. European Journal of Biochemistry, 268(23), 6011–6019. https://doi.org/10.1046/j.0014-2956.2001.02298.x.
Higashi, D. (1969). Purification and homogeneity of the histidine ammonia-lyase from Pseudomonas fluorescens. The Kumamoto Medical Journal, 22(4), 180–188.
Münch, A., Stingl, L., Jung, K., & Heermann, R. (2008). Photorhabdus luminescens genes induced upon insect infection. BMC Genomics, 9(1), 229. https://doi.org/10.1186/1471-2164-9-229.
Fischer-Le Saux, M., Viallard, V., Brunel, B., Normand, P., & Boemare, N. E. (1999). Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov. and P. asymbiotica sp. nov. International Journal of Systematic and Evolutionary Microbiology, 49(Pt 4), 1645–1656. https://doi.org/10.1099/00207713-49-4-1645.
Gatsogiannis, C., Lang, A. E., Meusch, D., Pfaumann, V., Hofnagel, O., Benz, R., Aktories, K., & Raunser, S. (2013). A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature, 495(7442), 520–523. https://doi.org/10.1038/nature11987.
Duchaud, E., Rusniok, C., Frangeul, L., Buchrieser, C., Givaudan, A., Taourit, S., Bocs, S., Boursaux-Eude, C., Chandler, M., Charles, J. F., Dassa, E., Derose, R., Derzelle, S., Freyssinet, G., Gaudriault, S., Médigue, C., Lanois, A., Powell, K., Siguier, P., Vincent, R., Wingate, V., Zouine, M., Glaser, P., Boemare, N., Danchin, A., & Kunst, F. (2003). The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nature Biotechnology, 21(11), 1307–1313. https://doi.org/10.1038/nbt886.
Lang, A. E., Schmidt, G., Schlosser, A., Hey, T. D., Larrinua, I. M., Sheets, J. J., Mannherz, H. G., & Aktories, K. (2010). Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science, 327(5969), 1139–1142. https://doi.org/10.1126/science.1184557.
Williams, J. S., Thomas, M., & Clarke, D. J. (2005). The gene stlA encodes a phenylalanine ammonia-lyase that is involved in the production of a stilbene antibiotic in Photorhabdus luminescens TT01. Microbiology, 151(Pt 8), 2543–2550. https://doi.org/10.1099/mic.0.28136-0.
Hyun, M. W., Yun, Y. H., Suh, D. Y., Han, J. H., & Kim, S. H. (2011). Immunological relationships among fungal and plant phenylalanine ammonia-lyases and bacterial histidine ammonia-lyase. The Korean Journal of Mycology, 39(3), 205–212. https://doi.org/10.4489/KJM.2010.39.3.205.
Okamura, H., Nishida, T., & Nakagawa, H. (1974). L-Histidine ammonia-lyase in rat liver. I. Purification and general characteristics. Journal of Biochemistry, 75(1), 139–152. https://doi.org/10.1093/oxfordjournals.jbchem.a130368.
Jun, S.-Y., Sattler, S. A., Cortez, G. S., Vermerris, W., Sattler, S. E., & Kang, C. (2018). Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase. Plant Physiology, 176(2), 1452–1468. https://doi.org/10.1104/pp.17.01608.
Hsieh, L. S., Ma, G. J., Yang, C. C., & Lee, P. D. (2010). Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry, 71(17-18), 1999–2009. https://doi.org/10.1016/j.phytochem.2010.09.019.
Xu, F., Deng, G., Cheng, S., Zhang, W., Huang, X., Li, L., Cheng, H., Rong, X., & Li, J. (2012). Molecular cloning, characterization and expression of the phenylalanine ammonia-lyase gene from Juglans regia. Molecules, 17(7), 7810–7823. https://doi.org/10.3390/molecules17077810.
Gao, J., Zhang, S., Feng, C., Zheng, X., Na, L., Qin, X., Ou, Y., Gu, X., Zhu, X., & Ying, X. (2012). Characterization, and expression profile of a phenylalanine ammonia lyase gene from Jatropha curcas L. Molecular Biology Reports, 39(4), 3443–3452. https://doi.org/10.1007/s11033-011-1116-4.
Bolwell, G. P., & Rodgers, M. W. (1991). L-Phenylalanine ammonia-lyase from French bean (Phaseolus vulgaris L.). Characterization and differential expression of antigenic multiple Mr forms. Biochemical Journal, 279(Pt 1), 231–236. https://doi.org/10.1042/bj2790231.
Wen, P.-F., Chen, J.-Y., Wan, S.-B., Kong, W.-F., Zhang, P., Wang, W., Zhan, J.-C., Pan, Q.-H., & Huang, W.-D. (2008). Salicylic acid activates phenylalanine ammonia-lyase in grape berry in response to high temperature stress. Plant Growth Regulation, 55(1), 1–10. https://doi.org/10.1007/s10725-007-9250-7.
Moffitt, M. C., Louie, G. V., Bowman, M. E., Pence, J., Noel, J. P., & Moore, B. S. (2007). Discovery of two cyanobacterial phenylalanine ammonia lyases: kinetic and structural characterization. Biochemistry, 46(4), 1004–1012. https://doi.org/10.1021/bi061774g.
Jia, S. R., Cui, J. D., Li, Y., & Sun, A. Y. (2008). Production of L-phenylalanine from trans-cinnamic acids by high-level expression of phenylalanine ammonia lyase gene from Rhodosporidium toruloides in Escherichia coli. Biochemical Engineering Journal, 42(3), 193–197. https://doi.org/10.1016/j.bej.2008.06.010.
El-Batal, A. I. (2002). Optimization of reaction conditions and stabilization of phenylalanine ammonia lyase-containing Rhodotorula glutinis cells during bioconversion of trans-cinnamic acid to L-phenylalanine. Acta Microbiologica Polonica, 51(2), 139–152.
Rösler, J., Krekel, F., Amrhein, N., & Schmid, J. (1997). Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiology, 113(1), 175–179. https://doi.org/10.1104/pp.113.1.175.
Jiang, H., Wood, K. V., & Morgan, J. A. (2005). Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Applied and Environmental Microbiology, 71(6), 2962–2969. https://doi.org/10.1128/AEM.71.6.2962-2969.2005.
Zang, Y., Jiang, T., Cong, Y., Zheng, Z., & Ouyang, J. (2015). Molecular characterization of a recombinant Zea mays phenylalanine ammonia-lyase (ZmPAL2) and its application in trans-cinnamic acid production from L-phenylalanine. Applied Biochemistry and Biotechnology, 176(3), 924–937. https://doi.org/10.1007/s12010-015-1620-4.
Cochrane, F. C., Davin, L. B., & Lewis, N. G. (2004). The Arabidopsis phenylalanine ammonia lyase gene family: kinetic characterization of the four PAL isoforms. Phytochemistry, 65(11), 1557–1564. https://doi.org/10.1016/j.phytochem.2004.05.006.
Cui, P., Zhong, W., Qin, Y., Tao, F., Wang, W., & Zhan, J. (2020). Characterization of two new aromatic amino acid lyases from actinomycetes for highly efficient production of p-coumaric acid. Bioprocess and Biosystems Engineering, 43(7), 1287–1298. https://doi.org/10.1007/s00449-020-02325-5.
Bazukian, I., Vardanian, A., Ambartsumian, A., Tozalakian, P., & IuG, P. (2009). Catalytic properties of Rhodotorula aurantiaca KM-1 phenylalanine ammonia-lyase. Applied Biochemistry and Microbiology, 45(1), 17–21. https://doi.org/10.1134/S0003683809010037.
Weise, N. J., Ahmed, S. T., Parmeggiani, F., Galman, J. L., Dunstan, M. S., Charnock, S. J., Leys, D., & Turner, N. J. (2017). Zymophore identification enables the discovery of novel phenylalanine ammonia lyase enzymes. Scientific Reports, 7(1), 13691. https://doi.org/10.1038/s41598-017-13990-0.
Xiang, L., & Moore, B. S. (2005). Biochemical characterization of a prokaryotic phenylalanine ammonia lyase. Journal of Bacteriology, 187(12), 4286–4289. https://doi.org/10.1128/JB.187.12.4286-4289.2005.
Author information
Authors and Affiliations
Contributions
JZ designed and coordinated the study. FZ and RJ performed the experiments. FZ, RJ, and JZ analyzed the results. FZ and JZ wrote the manuscript. All of the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
The research performed did not involve any human or animal subjects. The research did not require informed consent.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, F., Ren, J. & Zhan, J. Identification and Characterization of an Efficient Phenylalanine Ammonia-Lyase from Photorhabdus luminescens. Appl Biochem Biotechnol 193, 1099–1115 (2021). https://doi.org/10.1007/s12010-020-03477-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03477-6