Abstract
Astrocytes are glial cells that are abundant in the central nervous system (CNS) and that have important homeostatic and disease-promoting functions1. However, little is known about the homeostatic anti-inflammatory activities of astrocytes and their regulation. Here, using high-throughput flow cytometry screening, single-cell RNA sequencing and CRISPR–Cas9-based cell-specific in vivo genetic perturbations in mice, we identify a subset of astrocytes that expresses the lysosomal protein LAMP12 and the death receptor ligand TRAIL3. LAMP1+TRAIL+ astrocytes limit inflammation in the CNS by inducing T cell apoptosis through TRAIL–DR5 signalling. In homeostatic conditions, the expression of TRAIL in astrocytes is driven by interferon-γ (IFNγ) produced by meningeal natural killer (NK) cells, in which IFNγ expression is modulated by the gut microbiome. TRAIL expression in astrocytes is repressed by molecules produced by T cells and microglia in the context of inflammation. Altogether, we show that LAMP1+TRAIL+ astrocytes limit CNS inflammation by inducing T cell apoptosis, and that this astrocyte subset is maintained by meningeal IFNγ+ NK cells that are licensed by the microbiome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All raw and processed deep sequencing data have been deposited into GEO under the SuperSeries accession number GSE144859. Source data are provided with this paper.
References
Linnerbauer, M., Wheeler, M. A. & Quintana, F. J. Astrocyte crosstalk in CNS inflammation. Neuron 108, 608–622 (2020).
de Saint Basile, G., Ménasché, G. & Fischer, A. Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules. Nat. Rev. Immunol. 10, 568–579 (2010).
Aggarwal, B. B. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 3, 745–756 (2003).
Chao, C. C. et al. Metabolic control of astrocyte pathogenic activity via cPLA2-MAVS. Cell 179, 1483–1498 (2019).
Mächler, P. et al. In vivo evidence for a lactate gradient from astrocytes to neurons. Cell Metab. 23, 94–102 (2016).
Nagai, J. et al. Hyperactivity with disrupted attention by activation of an astrocyte synaptogenic cue. Cell 177, 1280–1292 (2019).
Allen, N. J. & Lyons, D. A. Glia as architects of central nervous system formation and function. Science 362, 181–185 (2018).
Sofroniew, M. V. Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 16, 249–263 (2015).
Alvarez, J. I. et al. The Hedgehog pathway promotes blood-brain barrier integrity and CNS immune quiescence. Science 334, 1727–1731 (2011).
Colonna, M. & Butovsky, O. Microglia function in the central nervous system during health and neurodegeneration. Annu. Rev. Immunol. 35, 441–468 (2017).
Liddelow, S. A. & Barres, B. A. Reactive astrocytes: production, function, and therapeutic potential. Immunity 46, 957–967 (2017).
Prinz, M., Jung, S. & Priller, J. Microglia biology: One century of evolving concepts. Cell 179, 292–311 (2019).
Wheeler, M. A. & Quintana, F. J. Regulation of astrocyte functions in multiple sclerosis. Cold Spring Harb. Perspect. Med. 9, a029009 (2019).
John Lin, C. C. et al. Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 20, 396–405 (2017).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Wheeler, M. A. et al. MAFG-driven astrocytes promote CNS inflammation. Nature 578, 593–599 (2020).
Ito, M. et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 565, 246–250 (2019).
Mayo, L. et al. IL-10-dependent Tr1 cells attenuate astrocyte activation and ameliorate chronic central nervous system inflammation. Brain 139, 1939–1957 (2016).
Kipnis, J. Multifaceted interactions between adaptive immunity and the central nervous system. Science 353, 766–771 (2016).
Rothhammer, V. et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016).
Erny, D. et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18, 965–977 (2015).
Rothhammer, V. et al. Microglial control of astrocytes in response to microbial metabolites. Nature 557, 724–728 (2018).
Sampson, T. R. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480 (2016).
Thion, M. S. et al. Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell 172, 500–516 (2018).
Wheeler, M. A. et al. Environmental control of astrocyte pathogenic activities in CNS inflammation. Cell 176, 581–596 (2019).
Pender, M. P., McCombe, P. A., Yoong, G. & Nguyen, K. B. Apoptosis of α β T lymphocytes in the nervous system in experimental autoimmune encephalomyelitis: its possible implications for recovery and acquired tolerance. J. Autoimmun. 5, 401–410 (1992).
Schmied, M. et al. Apoptosis of T lymphocytes in experimental autoimmune encephalomyelitis. Evidence for programmed cell death as a mechanism to control inflammation in the brain. Am. J. Pathol. 143, 446–452 (1993).
Zeine, R. & Owens, T. Loss rather than downregulation of CD4+ T cells as a mechanism for remission from experimental allergic encephalomyelitis. J. Neuroimmunol. 44, 193–198 (1993).
Lee, Y. et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012).
Ferber, I. A. et al. Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE). J. Immunol. 156, 5–7 (1996).
Wilson, N. S., Dixit, V. & Ashkenazi, A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat. Immunol. 10, 348–355 (2009).
Colonna, M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity 48, 1104–1117 (2018).
Filiano, A. J. et al. Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature 535, 425–429 (2016).
Ganal, S. C. et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity 37, 171–186 (2012).
Thiemann, S. et al. Enhancement of IFNγ production by distinct commensals ameliorates salmonella-induced disease. Cell Host Microbe 21, 682–694 (2017).
Tomura, M. et al. Monitoring cellular movement in vivo with photoconvertible fluorescence protein “Kaede” transgenic mice. Proc. Natl Acad. Sci. USA 105, 10871–10876 (2008).
Mayo, L. et al. Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nat. Med. 20, 1147–1156 (2014).
Bai, Z. et al. Cerebrospinal fluid and blood cytokines as biomarkers for multiple sclerosis: a systematic review and meta-analysis of 226 studies with 13,526 multiple sclerosis patients. Front. Neurosci. 13, 1026 (2019).
Foo, L. C. et al. Development of a method for the purification and culture of rodent astrocytes. Neuron 71, 799–811 (2011).
Baruch, K. et al. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 346, 89–93 (2014).
Gross, C. C. et al. Regulatory functions of natural killer cells in multiple sclerosis. Front. Immunol. 7, 606 (2016).
Gross, C. C. et al. Impaired NK-mediated regulation of T cell activity in multiple sclerosis is reconstituted by IL-2 receptor modulation. Proc. Natl Acad. Sci. USA 113, E2973–E2982 (2016).
Cella, M., Otero, K. & Colonna, M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1β reveals intrinsic functional plasticity. Proc. Natl Acad. Sci. USA 107, 10961–10966 (2010).
Sun, J. C., Beilke, J. N. & Lanier, L. L. Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009).
Ascherio, A., Munger, K. L. & Lünemann, J. D. The initiation and prevention of multiple sclerosis. Nat. Rev. Neurol. 8, 602–612 (2012).
Rojas, O. L. et al. Recirculating intestinal IgA-producing cells regulate neuroinflammation via IL-10. Cell 177, 492–493 (2019).
Kadowaki, A. & Quintana, F. J. The gut–CNS axis in multiple sclerosis. Trends Neurosci. 43, 622–634 (2020).
Korn, T. & Kallies, A. T cell responses in the central nervous system. Nat. Rev. Immunol. 17, 179–194 (2017).
Codarri, L. et al. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12, 560–567 (2011).
El-Behi, M. et al. The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12, 568–575 (2011).
Khakh, B. S. & Deneen, B. The emerging nature of astrocyte diversity. Annu. Rev. Neurosci. 42, 187–207 (2019).
Buffington, S. A. et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165, 1762–1775 (2016).
Chu, C. et al. The microbiota regulate neuronal function and fear extinction learning. Nature 574, 543–548 (2019).
Rothhammer, V. et al. Sphingosine 1-phosphate receptor modulation suppresses pathogenic astrocyte activation and chronic progressive CNS inflammation. Proc. Natl Acad. Sci. USA 114, 2012–2017 (2017).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Chen, S. et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 (2015).
Lee, Y., Messing, A., Su, M. & Brenner, M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia 56, 481–493 (2008).
Wang, T. et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168, 890–903 (2017).
Foo, L. C. Purification of rat and mouse astrocytes by immunopanning. Cold Spring Harb. Protoc. 2013, pdb.prot074211 (2013).
Beaudoin, G. M. III et al. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protoc. 7, 1741–1754 (2012).
Yates, A. et al. Ensembl 2016. Nucleic Acids Res. 44, D710–D716 (2016).
Ovcharenko, I. et al. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 15, 184–194 (2005).
Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015).
Xu, H. et al. Transcriptional atlas of intestinal immune cells reveals that neuropeptide α-CGRP modulates group 2 innate lymphoid cell responses. Immunity 51, 696–708 (2019).
Kim, R. Y. et al. Astrocyte CCL2 sustains immune cell infiltration in chronic experimental autoimmune encephalomyelitis. J. Neuroimmunol. 274, 53–61 (2014).
Schläger, C. et al. Effector T-cell trafficking between the leptomeninges and the cerebrospinal fluid. Nature 530, 349–353 (2016).
Kolev, M. et al. Diapedesis-induced integrin signaling via LFA-1 facilitates tissue immunity by inducing intrinsic complement C3 expression in immune cells. Immunity 52, 513–527 (2020).
Polman, C. H. et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 69, 292–302 (2011).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Zhang, X. et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell 73, 130–142 (2019).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Finak, G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Trombetta, J. J. et al. Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1–4.22.17 (2014).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019).
Acknowledgements
This work was supported by grants NS102807, ES02530, ES029136, AI126880 and AI149699 from the NIH; and RG4111A1 and JF2161-A-5 from the National Multiple Sclerosis Society (NMSS) (to F.J.Q.) F.J.Q. and A.P. received support from the International Progressive MS Alliance (PA-1604-08459); I.M.C. is supported by grants DP2AT009499 and R01AI130019 from the NIH, a Chan-Zuckerberg Initiative Ben Barres Early Career award and the Burroughs Wellcome fund; A.P. holds the T1 Canada Research Chair in MS and is funded by the Canada Institute of Health Research, the NMSS and the Canadian Foundation for Innovation; M.A.W. was supported by the NIH (1K99NS114111, F32NS101790), a training grant from the NIH and Dana-Farber Cancer Institute (T32CA207201), a travelling neuroscience fellowship from the Program in Interdisciplinary Neuroscience at the Brigham and Women’s Hospital and the Women’s Brain Initiative at the Brigham and Women’s Hospital; C.G.-V. was supported by an Alfonso Martín Escudero Foundation postdoctoral fellowship and by a postdoctoral fellowship from the European Molecular Biology Organization (ALTF 610-2017); and C.M.P. was supported by a fellowship from the FAPESP BEPE (2019/13731-0). Sanger sequencing was carried out at the DNA Resource Core of Dana-Farber/Harvard Cancer Center (funded in part by NCI Cancer Center support grant 2P30CA006516- 48). We thank T. Korn for sharing IfngEYFP reporter mice and U. von Andrian for sharing CAGKaede mice. We thank S. Boswell, M. Chatterjee, A. Ratner and the Harvard Medical School Single Cell Core for assistance with InDrop libraries; S. Garamszegi, T. Mason, E. LaRoche and the Broad Institute Genomics Platform for assistance with deep sequencing; R. Krishnan for assistance with flow cytometry studies, the use of the Harvard Digestive Disease Center (HDDC) core services, resources, technology and expertise; P. Hewson and G. Lipof for technical assistance; and all members of the Quintana laboratory for advice and discussions.
Author information
Authors and Affiliations
Contributions
L.M.S. and M.A.W. performed most in vivo, in vitro, FACS and genomics experiments. C.G.-V., C.M.P., M.L., F.G., G.S., E.S.H. and M.A. assisted with in vivo, in vitro, FACS and genomics experiments. F.A.P.-R. assisted with isolation and immunostaining of the meninges. L.M.S., M.A.W. and Z.L. performed bioinformatic analyses. K.V.B. performed RNAscope studies with L.M.S. and M.A.W. S.E.J.Z., D.L.R., B.B., I.M.C. and A.P. provided unique reagents and/or discussed and interpreted findings. L.M.S., M.A.W. and F.J.Q. wrote the manuscript with input from all co-authors. F.J.Q. designed and supervised the study and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Peter Calabresi, Hartmut Wekerle and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Profiling of astrocyte surface markers.
a, Sample FACS gating strategy. b, Histogram depicting the expression of surface markers in astrocytes.
Extended Data Fig. 2 Analysis of differentially expressed astrocyte surface markers.
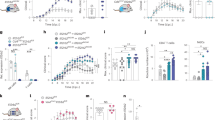
a, EAE in Aldh1l1creERT2tdTomato mice. n = 6 naive, n = 7 peak, n = 7 recovery mice. b, Sorting schematic for tdTomato+ astrocytes. c, MFI of LAMP1 expression from screening. n = 6 naive, n = 7 peak, n = 7 recovery mice. Unpaired two-tailed t-test. Data are mean ± s.e.m.
Extended Data Fig. 3 Analysis of LAMP1+ astrocytes.
a, Immunostaining of brain and spinal cord of mice transduced with Gfap-driven Cas9-2A-EGFP lentivirus. Representative images from n = 5 mice analysed. b, Percentage of GFP+GFAP+ cells by FACS. n = 5 mice. c, Quantification of astrocyte LAMP1 knockdown in astrocytes and microglia by FACS. n = 4 mice (sgScramble astrocyte); n = 5 mice otherwise. Unpaired two-tailed t-test. d, Gating strategy of CD4+ T cells from CNS. e, Analysis of splenic T cells from sgScramble- or sgLamp1-transduced mice. n = 5 mice sgLamp1 active-caspase 3; n = 6 mice otherwise. Unpaired two-tailed t-test. f, Analysis of CNS cells isolated from sgScramble- or sgLamp1-transduced mice. n = 6 per group. Unpaired two-tailed t-test. g, Analysis of CD11b+ cell apoptosis in sgScramble- or sgLamp1-transduced mice. n = 6 per group. Unpaired two-tailed t-test. h, Example gating used to sort LAMP1+ astrocytes from Aldh1l1creERT2tdTomato mice for scRNA-seq. i, Violin plots of genes, UMIs and cell-type markers for cells analysed by scRNA-seq from naive and EAE mice 17 days after induction. Astrocyte markers include Aqp4, Aldh1l1, Gfap, S100b, Aldoc, Slc1a3, Slc1a2 and Mfge8. j, Principal components used and gene expression scatterplots of LAMP1+ astrocytes isolated from Aldh1l1creERT2tdTomato mice. k, TRAIL expression on astrocytes from sgScramble- and sgLamp1-transduced mice 28 days after EAE induction. n = 5 sgScramble, n = 4 sgLamp1 mice. Unpaired two-tailed t-test. Data are mean ± s.e.m.
Extended Data Fig. 4 Analysis of TRAIL+ astrocytes.
a, Quantification of TRAIL knockdown using FACS in astrocytes and microglia. n = 5 mice per group. Unpaired two-tailed t-test. b, Analysis of CNS cell numbers in sgScramble- or sgTnfsf10-transduced mice. n = 6 mice per group. Unpaired two-tailed t-test. c, Control analyses of splenic T cells from knockdown mice. n = 6 mice per group. Unpaired two-tailed t-test. d, Analysis of CD11b+ cell apoptosis in sgScramble- or sgTnfsf10-transduced mice. n = 6 mice per group. Unpaired two-tailed t-test. e, Cytokine production and recall response of splenic T cells isolated from knockdown mice. n = 6 mice per group. n = 8 sgScramble CPM, n = 5 sgScramble IL-17 mice. Unpaired two-tailed t-test. f, Sorting schematic of LAMP1+TRAIL+ astrocytes from Aldh1l1creERT2tdTomato mice for scRNA-seq. g, Principal components used in scRNA-seq analysis of LAMP1+TRAIL+ astrocytes isolated from Aldh1l1creERT2tdTomato mice from naive and EAE mice at 17 days after induction. h, Violin plots of genes, UMIs and cell-type markers for cells analysed. Astrocyte markers include Aqp4, Aldh1l1, Gfap, S100b, Aldoc, Slc1a3, Slc1a2 and Mfge8. Data are mean ± s.e.m.
Extended Data Fig. 5 IFNγ–TRAIL signalling in other datasets.
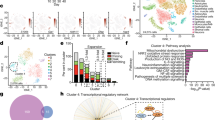
a, b, Identification of an IFNγ–TRAIL signalling network in a population of mouse astrocytes (cluster 6) previously analysed during the course of EAE in a previous study16. c, Comparison of the relative abundance of homeostatic vs pathogenic glia over the course of EAE. Compared to a pathogenic astrocyte population, astrocytes regulated by IFNγ and characterized by TRAIL are de-enriched during EAE. d, e, Identification of an IFNγ–TRAIL signalling network in a patient with MS or control astrocytes previously analysed by scRNA-seq during MS in a previous study16. n = 25 mice. n = 48 humans. f, Analysis by Drop-seq of tdTomato+ astrocytes isolated by flow cytometry. Data shown as mean ± s.e.m.
Extended Data Fig. 6 IFNγ signalling in LAMP1+TRAIL+ astrocytes.
a, STAT1 phosphorylation determined by flow cytometry in mouse astrocytes in culture treated with IFNγ or vehicle. n = 4 per condition. Unpaired two-tailed t-test. Experiment repeated three times. b, Analysis of Tnfsf10 expression in primary microglia and neurons in response to IFNγ. n = 8 for neuron IFNγ-treated, n = 5 neuron veh, n = 6 otherwise. Unpaired two-tailed t-test, data log-normalized for neuron Tnfsf10. c, Analysis of other apoptosis-promoting molecules expressed by astrocytes. n = 14 Fasl vehicle; n = 13 Gzmb vehicle; n = 12 Prf1 vehicle.; n = 7 Fasl IFNγ; n = 7 Prf1 IFNγ; n = 3 Gzmb IFNγ; n = 3 for Tnfsf10. Unpaired two-tailed t-test. Tnfsf10 data ln-transformed. Note the consistent axis scale. d, Analysis of Tnfsf10 expression by qPCR in astrocytes isolated from mice null for the indicated genes 29 days after EAE induction. n = 5 Ctrl, n = 3 Csf2rbf/f-Aldh1l1creERT2, n = 6 Tnfrsf1a−/−;Tnfrsf1b−/−mice. Unpaired two-tailed t-test (Tnfrsf1a−/−;Tnfrsf1b−/−) and Mann–Whitney test (Csf2rbf/f-Aldh1l1creERT2). e, EAE in Ifngr1+/+ (n = 12 mice) or Ifngr1−/− (n = 10 mice). Two-way repeated measures ANOVA. f, Flow cytometry analysis of T cells in the CNS of Ifngr1+/+ or Ifngr1−/− mice. n = 5 mice per group. Unpaired two-tailed t-test. g, Caspase-3 activation in CD4+ T cells isolated from the CNS of Ifngr1+/+ or Ifngr1−/− mice. n = 6 per group. Unpaired two-tailed t-test. Data are mean ± s.e.m.
Extended Data Fig. 7 Effects of astrocyte-specific knockdown of IFNγ signalling.
a, Knockdown analyses by FACS of mice transduced with sgScramble, sgIfngr1, sgIfngr2 and sgStat1. n = 5 mice per group. n = 4 sgScramble STAT1 mice. Unpaired two-tailed t-test. b, Control analyses of microglial expression following astrocyte knockdown. n = 5 sgIfngr2, n = 5 sgStat1, n = 6 mice otherwise. Unpaired two-tailed t-test. c, Analysis of Cxcl10 expression in astrocytes isolated from Ifngr1+/+, Ifngr1−/− or sgIfngr1 mice 29 days after EAE induction. n = 11 Ifngr1+/+, n = 6 Ifngr1−/−, n = 5 sgIfngr1. Unpaired two-tailed t-test. d, Analysis of astrocyte chemokine and T cell chemokine receptor expression from knockdown mice 29 days after EAE induction. n = 4 sgStat1, n = 4 sgIfngr2 Ccl20, n = 5 mice otherwise. Unpaired two-tailed t-test per condition. Data are mean ± s.e.m.
Extended Data Fig. 8 Microglia monocyte, and peripheral T cell responses to astrocyte perturbation.
a–d, Bulk RNA-seq analyses of microglia (CD11b+CD45+Ly6Clow) isolated by flow cytometry from mice transduced with the indicated astrocyte-targeting CRISPR–Cas9 lentivirus 24 days after EAE induction. Heat map of differentially expressed genes, GSEA analysis, Qiagen IPA and volcano plot shown per group. n = 4 mice per group, n = 3 mice for sgTnfsf10. e, Control analyses of CNS-recruited pro-inflammatory monocytes. n = 6 sgScramble, sgIfngr1; n = 5 mice otherwise. One-way ANOVA, Sidak post-hoc test. f, Control analyses of splenic T cells. n = 6 sgScramble, sgIfngr1; n = 5 sgIfngr1 CD4+, n = 5 mice otherwise. One-way ANOVA, Dunnett post-hoc test. g, Analysis of active-caspase 3 expression in CD11b+ cells. n = 6 sgScramble, sgIfngr1; n = 5 mice otherwise. One-way ANOVA, Dunnett post-hoc test. h, Cytokine production and recall response of splenic T cells isolated from knockdown mice. n = 6 per group. n = 5 sgScramble IL-17, n = 8 sgScramble, sgIfngr1, sgStat1 CPM mice. One-way ANOVA, Dunnett post-hoc test. Data are mean ± s.e.m.
Extended Data Fig. 9 Control analyses of in vitro apoptosis and anti-NK1.1 antibody depletion.
a, Active caspase-3 and TUNEL FACS analysis in CD4+ T cells co-cultured with astrocytes pre-treated with vehicle or IFNγ. n = 3 for active-caspase 3, n = 2 for TUNEL vehicle, n = 4 for TUNEL IFNγ. Unpaired two-tailed t-test. b, Active caspase-3 expression in CD4+ T cells co-cultured with astrocytes transfected with the indicated siRNA. n = 4 per condition. Unpaired two-tailed t-test. c, Left, DR5 expression in cultured IFNγ+, IFNγ+IL-17+ and IFNγ-IL-17− CD4+ T cells activated in vitro. n = 3 per group. One-way ANOVA, Dunnett post-hoc test. Right, active-caspase 3 expression in DR5+ or DR5− CD4+ T cells co-cultured with IFNγ-pre-treated astrocytes. n = 3 per group. Unpaired two-tailed t-test. d, Negative control images of IFNγ immunostaining using Ifng−/− mice. Repeated twice. e, Fluorescent in situ hybridization (FISH) of Gfap co-stained with anti-GFAP antibody. Repeated three times. f, FISH of Tnfsf10 and Ifng in naive or EAE mouse spinal cord co-stained with anti-GFAP antibody. n = 8 images (naive), n = 6 images (EAE). Unpaired two-tailed t-test. Repeated three times. g, FISH of Ifng, Tnfsf10, and Klrb1c in naive or EAE mouse spinal cord. Green arrowhead, NK cell; red arrowhead, astrocyte. Repeated twice. h, Control analyses for anti-NK1.1 antibody depletion experiment. n = 4 per group. Unpaired two-tailed t-test. i, Quantification of splenic NKp46+ cells during α-NK1.1 antibody depletion experiment. n = 4 per group. Unpaired two-tailed t-test. j, Quantification of NK1.1+CD200R- NK cells in the spleen during α-NK1.1 antibody depletion experiment. n = 4 per group. Unpaired two-tailed t-test. Data are mean ± s.e.m.
Extended Data Fig. 10 Control analyses of microbial regulation of NK cells and astrocytes.
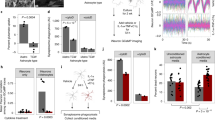
a, Quantification of astrocytes from SPF (n = 3) or GF (n = 5) mice. Unpaired two-tailed t-test. b, Quantification of CD4+ T cells in SPF and GF mice. Splenic T cells: n = 4 per group; lamina propria T cells: n = 3 per group. Unpaired two-tailed t-test. c, IFNγ+ NK cells in spleen and lamina propria of SPF and GF mice. n = 5 per group (spleen), n = 6 per group (lamina propria). Unpaired two-tailed t-test. d, Sorting schematic (left) and splenic cell numbers (right) from donors and recipients in NK cell adoptive transfer experiments. Quantification left-to-right: glow cytometry quantification of splenic IFNγ+ NK cells from donor mice. n = 5 GF, n = 8 SPF WT, n = 7 SPF Ifng−/− mice. Total number of NK cells from donor mice. n = 6 GF, n = 8 mice otherwise. Unpaired two-tailed t-test. Total number of NK cells in recipient spleens: n = 4 mice per group. One-way ANOVA, Tukey’s post-hoc test. e, Analysis by qPCR of 16S rRNA abundance as a function of two-week antibiotic treatment (ABX) and ABX treatment followed by faecal microbiota transplant (FMT) in IfngEYFP mice. n = 6 mice per group. Unpaired two-tailed t-test. f, Gating schematic for the meninges of IfngEYFP mice. g, FACS plots of IFNγ expression during three-week antibiotic depletion of commensal bacteria (ABX) or two-week ABX followed by 1-week reconstitution by faecal microbiota transplant (FMT) using IfngEYFP reporter mice. h, Analysis of NK cells in the spleen and lamina propria as a function of ABX and ABX + FMT treatment using IfngEYFP reporter mice. n = 6 mice for total NK cells ABX and ABX + FMT, n = 10 mice for vehicle meninges condition, n = 5 mice otherwise. Unpaired two-tailed t-test. i, Gating schematic of NK cell sorting from the spleen of CAGKaede mice. j, Analysis by qPCR of bacterial 16S rRNA abundance as a function of ABX and ABX + FMT in CAGKaede mice. n = 9 mice per group. Unpaired two-tailed t-test. Data are mean ± s.e.m.
Supplementary information
Supplementary Table 1
Results from astrocyte surface marker screen.
Supplementary Table 2
Differential gene expression of astrocytes analyzed by RNA-seq isolated from sgScrmbl- versus sgLamp1-knockdown mice during EAE. P-value determined by MAST algorithm.
Supplementary Table 3
Differential gene expression by cluster of LAMP1+ astrocytes isolated from AldhCreERT2tdTomato mice and analyzed by inDrop scRNA-seq. FDR determined by Benjamini-Hochberg test.
Supplementary Table 4
Differential gene expression of astrocytes analyzed by RNA-seq isolated from sgScrmbl- versus sgTnfsf10-knockdown mice during EAE. P-value determined by MAST algorithm.
Supplementary Table 5
Differential gene expression by cluster of LAMP1+TRAIL+ astrocytes isolated from AldhCreERT2tdTomato mice and analyzed by inDrop scRNA-seq. FDR determined by Benjamini-Hochberg test.
Supplementary Table 6
Differential gene expression by cluster of tdTomato+ astrocytes analyzed by Drop-seq. FDR determined by Benjamini-Hochberg test.
Supplementary Table 7
Differential gene expression of astrocytes analyzed by RNA-seq isolated from sgScrmbl- versus sgIfngr1-, sgIfngr2-, and sgStat1-knockdown mice during EAE. P-value determined by MAST algorithm.
Source data
Rights and permissions
About this article
Cite this article
Sanmarco, L.M., Wheeler, M.A., Gutiérrez-Vázquez, C. et al. Gut-licensed IFNγ+ NK cells drive LAMP1+TRAIL+ anti-inflammatory astrocytes. Nature 590, 473–479 (2021). https://doi.org/10.1038/s41586-020-03116-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-03116-4
This article is cited by
-
Understanding immune microenvironment alterations in the brain to improve the diagnosis and treatment of diverse brain diseases
Cell Communication and Signaling (2024)
-
Regulation of stress granule formation in human oligodendrocytes
Nature Communications (2024)
-
Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases
Signal Transduction and Targeted Therapy (2024)
-
Disease-associated astrocyte epigenetic memory promotes CNS pathology
Nature (2024)
-
The astrocyte-produced growth factor HB-EGF limits autoimmune CNS pathology
Nature Immunology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.