Abstract
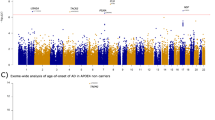
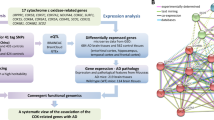
Accumulating evidence demonstrated that GABAergic dysfunction contributes to the pathogenesis of Alzheimer’s disease (AD). The GABA aminotransferase (ABAT) gene encodes a mitochondrial GABA transaminase and plays key roles in the biogenesis and metabolism of gamma-aminobutyric acid (GABA), which is a major inhibitory neurotransmitter. In this study, we performed an integrative study at the genetic and expression levels to investigate the potential genetic association between the ABAT gene and AD. Through re-analyzing data from the currently largest meta-analysis of AD genome-wide association study (GWAS), we identified genetic variants in the 3’-UTR of ABAT as the top AD-associated SNPs (P < 1 × 10−4) in this gene. Functional annotation of these AD-associated SNPs indicated that these SNPs are located in the regulatory regions of transcription factors or/and microRNAs. Expression quantitative trait loci (eQTL) analysis and luciferase reporter assay showed that the AD risk alleles of these SNPs were associated with a reduced expression level of ABAT. Further analysis of mRNA expression data and single-cell transcriptome data of AD patients showed that ABAT reduction in the neuron is an early event during AD development. Overall, our results indicated that ABAT genetic variants may be associated with AD through affecting its mRNA expression. An abnormal level of ABAT will lead to a disturbance of the GABAergic signal pathway in AD brains.



Similar content being viewed by others
Change history
22 January 2021
A Correction to this paper has been published: https://doi.org/10.1007/s12035-021-02301-4
References
Alzheimer’s Association (2020) 2020 Alzheimer’s disease facts and figures. Alzheimers Dement 16:391–460
Huang Y, Mucke L (2012) Alzheimer mechanisms and therapeutic strategies. Cell 148(6):1204–1222
Querfurth HW, LaFerla FM (2010) Alzheimer’s disease. N Engl J Med 362(4):329–344
De Strooper B, Karran E (2016) The cellular phase of Alzheimer’s disease. Cell 164(4):603–615
Sims R, Hill M, Williams J (2020) The multiplex model of the genetics of Alzheimer’s disease. Nat Neurosci 23(3):311–322
Karch CM, Goate AM (2015) Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 77(1):43–51
Zhang DF, Xu M, Bi R, Yao YG (2019) Genetic analyses of Alzheimer’s disease in China: achievements and perspectives. ACS Chem Neurosci 10(2):890–901
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al (2013)Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12):1452–1458
Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, Sealock J, Karlsson IK et al (2019)Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 51(3):404–413
Bis JC, Jian X, Kunkle BW, Chen Y, Hamilton-Nelson KL, Bush WS et al (2020) Whole exome sequencing study identifies novel rare and common Alzheimer’s-associated variants involved in immune response and transcriptional regulation. Mol Psychiatry 25(8):1859–1875
Zhang DF, Fan Y, Xu M, Wang G, Wang D, Li J, Kong LL, Zhou H et al (2019) Complement C7 is a novel risk gene for Alzheimer’s disease in Han Chinese. Natl Sci Rev 6(2):257–274
Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J et al (2013) Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368(2):107–116
Zhou X, Chen Y, Mok KY, Zhao Q, Chen K, Hardy J et al (2018) Identification of genetic risk factors in the Chinese population implicates a role of immune system in Alzheimer’s disease pathogenesis. Proc Natl Acad Sci U S A 115(8):1697–1706
Ridge PG, Mukherjee S, Crane PK, Kauwe JS (2013) Alzheimer’s disease: analyzing the missing heritability. PLoS One 8(11):e79771
Palop JJ, Mucke L (2016) Network abnormalities and interneuron dysfunction in Alzheimer disease. Nat Rev Neurosci 17(12):777–792
Bi D, Wen L, Wu Z, Shen Y (2020) GABAergic dysfunction in excitatory and inhibitory (E/I) imbalance drives the pathogenesis of Alzheimer’s disease. Alzheimers Dement 16(9):1312–1329
Owens DF, Kriegstein AR (2002) Is there more to GABA than synaptic inhibition? Nat Rev Neurosci 3(9):715–727
Chen WF, Maguire S, Sowcik M, Luo W, Koh K, Sehgal A (2015) A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol Psychiatry 20(2):240–251
Prevot T, Sibille E (2020) Altered GABA-mediated information processing and cognitive dysfunctions in depression and other brain disorders. Mol Psychiatry. https://doi.org/10.1038/s41380-020-0727-3
Martin-Belmonte A, Aguado C, Alfaro-Ruiz R, Moreno-Martinez AE, de la Ossa L, Martinez-Hernandez J et al (2020) Density of GABAB receptors is reduced in granule cells of the hippocampus in a mouse model of Alzheimer’s disease. Int J Mol Sci 21(7)
Martin-Belmonte A, Aguado C, Alfaro-Ruiz R, Moreno-Martinez AE, de la Ossa L, Martinez-Hernandez J et al (2020) Reduction in the neuronal surface of post and presynaptic GABAB receptors in the hippocampus in a mouse model of Alzheimer’s disease. Brain Pathol 30(3):554–575
Limon A, Reyes-Ruiz JM, Miledi R (2012) Loss of functional GABA(A) receptors in the Alzheimer diseased brain. Proc Natl Acad Sci U S A 109(25):10071–10076
Garcia-Marin V, Blazquez-Llorca L, Rodriguez JR, Boluda S, Muntane G, Ferrer I et al (2009) Diminished perisomatic GABAergic terminals on cortical neurons adjacent to amyloid plaques. Front Neuroanat 3:28
Brawek B, Chesters R, Klement D, Muller J, Lerdkrai C, Hermes M et al (2018) A bell-shaped dependence between amyloidosis and GABA accumulation in astrocytes in a mouse model of Alzheimer’s disease. Neurobiol Aging 61:187–197
Park JH, Ju YH, Choi JW, Song HJ, Jang BK, Woo J et al (2019) Newly developed reversible MAO-B inhibitor circumvents the shortcomings of irreversible inhibitors in Alzheimer’s disease. Sci Adv 5(3):eaav0316
Lu MH, Zhao XY, Xu DE, Chen JB, Ji WL, Huang ZP, Pan TT, Xue LL et al (2020) Transplantation of GABAergic interneuron progenitor attenuates cognitive deficits of Alzheimer’s disease model mice. J Alzheimers Dis 75(1):245–260
Govindpani K, Calvo-Flores Guzman B, Vinnakota C, Waldvogel HJ, Faull RL, Kwakowsky A (2017) Towards a better understanding of GABAergic remodeling in Alzheimer’s disease. Int J Mol Sci 18(8)
Najm R, Jones EA, Huang Y (2019) Apolipoprotein E4, inhibitory network dysfunction, and Alzheimer’s disease. Mol Neurodegener 14(1):24
Rice HC, de Malmazet D, Schreurs A, Frere S, Van Molle I, Volkov AN et al (2019) Secreted amyloid-beta precursor protein functions as a GABABR1a ligand to modulate synaptic transmission. Science 363(6423):eaao4827
Calvo-Flores Guzman B, Kim S, Chawdhary B, Peppercorn K, Tate WP, Waldvogel HJ et al (2020)Amyloid-Beta1-42-induced increase in GABAergic tonic conductance in mouse hippocampal CA1 pyramidal cells. Molecules 25(3):693
Zheng J, Li HL, Tian N, Liu F, Wang L, Yin Y, Yue L, Ma L et al (2020) Interneuron accumulation of phosphorylated tau impairs adult hippocampal neurogenesis by suppressing GABAergic transmission. Cell Stem Cell 26(3):331–345 e336
Sherif FM, Ahmed SS (1995) Basic aspects of GABA-transaminase in neuropsychiatric disorders. Clin Biochem 28(2):145–154
Ambrad Giovannetti E, Fuhrmann M (2019) Unsupervised excitation: GABAergic dysfunctions in Alzheimer’s disease. Brain Res 1707:216–226
Koenig MK, Hodgeman R, Riviello JJ, Chung W, Bain J, Chiriboga CA, Ichikawa K, Osaka H et al (2017) Phenotype of GABA-transaminase deficiency. Neurology 88(20):1919–1924
Aoyagi T, Wada T, Nagai M, Kojima F, Harada S, Takeuchi T et al (1990) Increased gamma-aminobutyrate aminotransferase activity in brain of patients with Alzheimer’s disease. Chem Pharm Bull (Tokyo) 38(6):1748–1749
Aoyagi T, Wada T, Kojima F, Nagai M, Harada S, Takeuchi T et al (1990) Increase in aminobutyrate aminotransferase and cholineacetyltransferase in cerebrum of aged rats. Chem Pharm Bull (Tokyo) 38(6):1750–1752
Sherif F, Gottfries CG, Alafuzoff I, Oreland L (1992) Brain gamma-aminobutyrate aminotransferase (GABA-T) and monoamine oxidase (MAO) in patients with Alzheimer’s disease. J Neural Transm Park Dis Dement Sect 4(3):227–240
Hooli BV, Kovacs-Vajna ZM, Mullin K, Blumenthal MA, Mattheisen M, Zhang C, Lange C, Mohapatra G et al (2014) Rare autosomal copy number variations in early-onset familial Alzheimer’s disease. Mol Psychiatry 19(6):676–681
Beecham GW, Bis JC, Martin ER, Choi SH, DeStefano AL, van Duijn CM et al (2017) The Alzheimer’s disease sequencing project: study design and sample selection. Neurol Genet 3(5):e194
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P et al (2015) UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12(3):e1001779
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR et al (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26(18):2336–2337
Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K et al (2018) The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46(D1):D794–D801
Chen Y, Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48(D1):D127–D131
Liu W, Wang X (2019) Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol 20(1):18
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46(3):310–315
Qi T, Wu Y, Zeng J, Zhang F, Xue A, Jiang L et al (2018) Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun 9(1):2282
GTEx Consortium (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45(6):580–585
GTEx Consortium (2015) Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348(6235):648–660
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC et al (2016) Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19(11):1442–1453
Ng B, White CC, Klein HU, Sieberts SK, McCabe C, Patrick E, Xu J, Yu L et al (2017) An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci 20(10):1418–1426
Xu M, Zhang DF, Luo R, Wu Y, Zhou H, Kong LL, Bi R, Yao YG (2018) A systematic integrated analysis of brain expression profiles reveals YAP1 and other prioritized hub genes as important upstream regulators in Alzheimer’s disease. Alzheimers Dement 14(2):215–229
Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L et al (2019)Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570(7761):332–337
Bam M, Yang X, Busbee BP, Aiello AE, Uddin M, Ginsberg JP, Galea S, Nagarkatti PS et al (2020) Increased H3K4me3 methylation and decreased miR-7113-5p expression lead to enhanced Wnt/beta-catenin signaling in immune cells from PTSD patients leading to inflammatory phenotype. Mol Med 26(1):110
Wegerer M, Adena S, Pfennig A, Czamara D, Sailer U, Bettecken T, Müller-Myhsok B, Modell S et al (2013) Variants within the GABA transaminase (ABAT) gene region are associated with somatosensory evoked EEG potentials in families at high risk for affective disorders. Psychol Med 43(6):1207–1217
Besse A, Wu P, Bruni F, Donti T, Graham BH, Craigen WJ, McFarland R, Moretti P et al (2015) The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism. Cell Metab 21(3):417–427
Devine MJ, Kittler JT (2018) Mitochondria at the neuronal presynapse in health and disease. Nat Rev Neurosci 19(2):63–80
Mattson MP, Gleichmann M, Cheng A (2008) Mitochondria in neuroplasticity and neurological disorders. Neuron 60(5):748–766
Availability of Data and Material
All data generated or analyzed in this study are included in the current manuscript and supplementary materials.
Funding
This study was supported by the National Natural Science Foundation of China (31730037, 31970560 and 31970965), Yunnan Province (2019FA027), the Strategic Priority Research Program (B) of the Chinese Academy of Sciences (CAS) (XDB02020003), the Key Research Program of Frontier Sciences, CAS (QYZDJ-SSW-SMC005), and the International Partnership Program of CAS (152453KYSB20170031).
Author information
Authors and Affiliations
Contributions
Y.-G.Y. and R.B. designed the study; M.X. and D.-F.Z. analyzed the data; R.B. and Q.Z. performed the experiments; R.B., Q.Z., and Y.-G.Y. drafted the manuscript; all authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors have seen and approved the manuscript and contributed significantly to this work.
Code Availability
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Assignment of affiliation number to “Yong-Gang Yao” should be 2, 3, 4.
Rights and permissions
About this article
Cite this article
Zheng, Q., Bi, R., Xu, M. et al. Exploring the Genetic Association of the ABAT Gene with Alzheimer’s Disease. Mol Neurobiol 58, 1894–1903 (2021). https://doi.org/10.1007/s12035-020-02271-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-020-02271-z