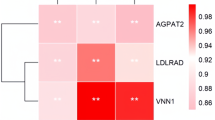
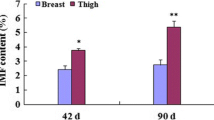
Abstract
Nandan-Yao chicken is a Chinese native chicken with lower fat deposition and better meat quality. Fat deposition is a quite complex and important economic trait. However, its molecular mechanism is still unknown in chickens. In the current study, Nandan-Yao chicken was divided into two groups based on the rate of abdominal fat at 120 days old, namely the high-fat group and low-fat group. The total RNAs were isolated and sequenced by RNA sequencing (RNA-seq). After quality control, we gained 1222, 902, 784, 624, and 736 differentially expressed genes (DEGs) in abdominal fat, back skin, liver, pectoral muscle, and leg muscle, respectively. Analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) showed that significantly enriched GO term and KEGG signaling pathway mainly involved cytosolic ribosome, growth development, PPAR signaling pathway, Wnt signaling pathway, and linoleic acid metabolism in abdominal fat, back skin, and liver. While in pectoral muscle and leg muscle, it is mainly enriched in phosphatidylinositol signaling system, adrenergic signaling in cardiomyocytes, cytosolic ribosome, and cytosolic part. Sixteen genes were differentially expressed in all five tissues. Among them, PLA2G4A and RPS4Y1 might be the key regulators for fat deposition in Nandan-Yao chicken. The protein-protein interaction (PPI) network analysis of DEGs showed that PCK1 was the most notable genes. The findings in the current study will help to understand the regulation mechanism of abdominal fat and intramuscular fat in Nandan-Yao chicken and provide a theoretical basis for Chinese local chicken breeding.








Similar content being viewed by others
Abbreviations
- RNA-seq:
-
RNA sequencing
- QPCR:
-
quantitative real-time PCR
- DEG:
-
differentially expressed genes
- PPI:
-
protein-protein interaction
- FPKM:
-
fragments per kilobase of transcript per million mapped reads
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
References
Aggrey SE, Karnuah AB, Sebastian B, Anthony NB (2010) Genetic properties of feed efficiency parameters in meat-type chickens. Genet Sel Evol 42:25. https://doi.org/10.1186/1297-9686-42-25
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics UBJ 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Abdalla BA, Chen J, Nie Q, Zhang X (2018) Genomic insights into the multiple factors controlling abdominal fat deposition in a chicken model. Genetics 9:262. https://doi.org/10.3389/fgene.2018.00262
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2. https://doi.org/10.1186/1471-2105-4-2
Beale EG, Forest C, Hammer RE (2003) Regulation of cytosolic phosphoenolpyruvate carboxykinase gene expression in adipocytes. Biochimie 85:1207–1211. https://doi.org/10.1016/j.biochi.2003.10.012
Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, Harrison SD, MacDougald OA (2002) Regulation of Wnt signaling during adipogenesis. J Biol Chem 277:30998–31004. https://doi.org/10.1074/jbc.M204527200
Cao X et al (2018) Integrating CNVs into meta-QTL identified GBP4 as positional candidate for adult cattle stature. Funct Integr Genomics 18:559–567. https://doi.org/10.1007/s10142-018-0613-0
Christodoulides C, Lagathu C, Sethi JK, Vidal-Puig A (2009) Adipogenesis and WNT signalling. Trends Endocrinol Metab 20:16–24. https://doi.org/10.1016/j.tem.2008.09.002
Cui HX, Liu RR, Zhao GP, Zheng MQ, Chen JL, Wen J (2012) Identification of differentially expressed genes and pathways for intramuscular fat deposition in pectoralis major tissues of fast-and slow-growing chickens. BMC Genomics 13:213. https://doi.org/10.1186/1471-2164-13-213
Droval AA, Benassi VT, Rossa A, Prudencio SH, Paiao FG, Shimokomaki M (2012) Consumer attitudes and preferences regarding pale, soft, and exudative broiler breast meat. J Appl Poult Res 21:502–507. https://doi.org/10.3382/japr.2011-00392
Duan J et al (2013) Androgen inhibits abdominal fat accumulation and negatively regulates the PCK1 gene in male chickens. PLoS One 8:e59636. https://doi.org/10.1371/journal.pone.0059636
Dunnington EA, Siegel PB (1996) Long-term divergent selection for eight-week body weight in white Plymouth rock chickens. Poult Sci 75:1168–1179. https://doi.org/10.3382/ps.0751168
Excel 2013 (Microsoft Corporation, Redmond, Washington, USA)
Forest C et al (2003) Fatty acid recycling in adipocytes: a role for glyceroneogenesis and phosphoenolpyruvate carboxykinase. Biochem Soc Trans 31:1125–1129. https://doi.org/10.1042/bst0311125
Guixian W, Qi Y, Liu X, Yang N, Xu G, Liu L, Li X (2017) Cecal MicroRNAome response to Salmonella enterica serovar Enteritidis infection in White Leghorn Layer. BMC Genomics 18:77. https://doi.org/10.1186/s12864-016-3413-8
Guangchuang Y, Wang L-G, Han Y, He Q-Y (2012) ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics: a Journal of Integrative Biology 16:284–287. https://doi.org/10.1089/omi.2011.0118
Hakimi P, Yang J, Casadesus G, Massillon D, Tolentino-Silva F, Nye CK, Cabrera ME, Hagen DR, Utter CB, Baghdy Y, Johnson DH, Wilson DL, Kirwan JP, Kalhan SC, Hanson RW (2007) Overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) in skeletal muscle repatterns energy metabolism in the mouse. J Biol Chem 282:32844–32855. https://doi.org/10.1074/jbc.M706127200
Hocquette JF, Gondret F, Baeza E, Medale F, Jurie C, Pethick DW (2010) Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers. Animal 4:303–319. https://doi.org/10.1017/S1751731109991091
Invernizzi G, Thering BJ, McGuire MA, Savoini G, Loor JJ (2010) Sustained upregulation of stearoyl-CoA desaturase in bovine mammary tissue with contrasting changes in milk fat synthesis and lipogenic gene networks caused by lipid supplements. Funct Integr Genomics 10:561–575. https://doi.org/10.1007/s10142-010-0179-y
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Liu L, Cui HX, Zheng MQ, Zhao GP, Wen J (2018a) Comparative analysis of differentially expressed genes related to triglyceride metabolism between intramuscular fat and abdominal fat in broilers. Br Poult Sci 59:514–520. https://doi.org/10.1080/00071668.2018.1483573
Li D, Pan Z, Zhang K, Yu M, Yu D, Lu Y, Wang J, Zhang J, Zhang K, du W (2020) Identification of the differentially expressed genes of muscle growth and intramuscular fat metabolism in the development stage of yellow broilers. Genes (Basel) 11(3):244. https://doi.org/10.3390/genes11030244
Li FG, Hu G, Zhang H, Wang SZ, Wang ZP, Li H (2013) Epistatic effects on abdominal fat content in chickens: results from a genome-wide SNP-SNP interaction analysis. PLoS One 8:9. https://doi.org/10.1371/journal.pone.0081520
Li H, Deeb N, Zhou H, Mitchell AD, Ashwell CM, Lamont SJ (2003) Chicken quantitative trait loci for growth and body composition associated with transforming growth factor-beta genes. Poult Sci 82:347–356. https://doi.org/10.1093/ps/82.3.347
Li H, Wang T, Xu C, Wang D, Ren J, Li Y, Tian Y, Wang Y, Jiao Y, Kang X, Liu X (2015) Transcriptome profile of liver at different physiological stages reveals potential mode for lipid metabolism in laying hens. BMC Genomics 16:13. https://doi.org/10.1186/s12864-015-1943-0
Liu RR, Sun Y, Zhao G, Wang F, Wu D, Zheng M, Chen J, Zhang L, Hu Y, Wen J (2013) Genome-wide association study identifies loci and candidate genes for body composition and meat quality traits in Beijing-You chickens. PLoS One 8:11. https://doi.org/10.1371/journal.pone.0061172
Liu Z, Meng J, Li X, Zhu F, Liu T, Wu G, Zhang L (2018b) Identification of hub genes and key pathways associated with two subtypes of diffuse large B-cell lymphoma based on gene expression profiling via integrated bioinformatics. Biomed Res Int 2018:3574534. https://doi.org/10.1155/2018/3574534
Longo KA, Wright WS, Kang S, Gerin I, Chiang SH, Lucas PC, Opp MR, MacDougald OA (2004) Wnt10b inhibits development of white and brown adipose tissues. J Biol Chem 279:35503–35509. https://doi.org/10.1074/jbc.M402937200
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. https://doi.org/10.1038/nmeth.1226
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Pang CY, Bai MZ, Zhang C, Chen J, Lu XR, Deng TX, Ma XY, Duan AQ, Liang SS, Huang YQ, Xiu Z, Liang XW (2019) Global transcriptome analysis of different stages of preimplantation embryo development in river buffalo. PeerJ 7:e8185. https://doi.org/10.7717/peerj.8185
Patience JF, Rossoni-Serao MC, Gutierrez NA (2015) A review of feed efficiency in swine: biology and application. J Anim Sci Biotechnol 6:33. https://doi.org/10.1186/s40104-015-0031-2
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT. StringTie and Ballgown Nat Protoc 11:1650–1667. https://doi.org/10.1038/nprot.2016.095
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295. https://doi.org/10.1038/nbt.3122
Qiangchuan H, Kwok L-Y, Zheng Y, Wang L et al (2016) Differential fecal microbiota are retained in broiler chicken lines divergently selected for fatness traits. Sci Rep 6:37376. https://doi.org/10.1038/srep37376
Resnyk CW, Chen C, Huang H, Wu CH, Simon J, le Bihan-Duval E, Duclos MJ, Cogburn LA (2015) RNA-Seq analysis of abdominal fat in genetically fat and lean chickens highlights a divergence in expression of genes controlling adiposity, Hemostasis, and Lipid Metabolism. PLoS One 10:41. https://doi.org/10.1371/journal.pone.0139549
Sahraeian SME, Mohiyuddin M, Sebra R, Tilgner H, Afshar PT, Au KF, Bani Asadi N, Gerstein MB, Wong WH, Snyder MP, Schadt E, Lam HYK (2017) Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun 8:15. https://doi.org/10.1038/s41467-017-00050-4
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. https://doi.org/10.1093/bioinformatics/btr026
Semakova J, Hyroššová P, Méndez-Lucas A, Cutz E, Bermudez J, Burgess S, Alcántara S, Perales JC (2017) PEPCK-C reexpression in the liver counters neonatal hypoglycemia in Pck1 (del/del) mice, unmasking role in non-gluconeogenic tissues. J Physiol Biochem 73:89–98. https://doi.org/10.1007/s13105-016-0528-y
Tan S, Reverter A, Wang Y, Byrne K, McWilliam S, Lehnert SJF (2006) Gene expression profiling of bovine in vitro adipogenesis using a cDNA microarray. Funct Integr Genomics 6:235–249. https://doi.org/10.1007/s10142-005-0016-x
Willson NL, Forder REA, Tearle R, Williams JL, Hughes RJ, Nattrass GS, Hynd PI (2018) Transcriptional analysis of liver from chickens with fast (meat bird), moderate (F1 layer x meat bird cross) and low (layer bird) growth potential. BMC Genomics 19:309. https://doi.org/10.1186/s12864-018-4723-9
Wright WS et al (2007) Wnt10b inhibits obesity in ob/ob and agouti mice. Diabetes 56:295–303. https://doi.org/10.2337/db06-1339
Xiaodan Q, Zeng D, Wang H et al (2018) Analysis of hepatic transcriptome demonstrates altered lipid metabolism following Lactobacillus johnsonii BS15 prevention in chickens with subclinical necrotic enteritis. Lipids Health Dis 17:93. https://doi.org/10.1186/s12944-018-0741-5
Xing K, Qi X, Ni H, Wang X, Guo Y, Sheng X (2019) Transcriptional analyses of endometrial caruncles in sheep with in vivo / in vitro produced embryos during the peri-implantation period. Reprod Biol 19:404–411. https://doi.org/10.1016/j.repbio.2019.09.008
Yuying L, Mo G, Sun J, Wei F, Liao DJ (2016) Genetic diversity of Guangxi chicken breeds assessed with microsatellites and the mitochondrial DNA D-loop region. Mol Biol Rep 43:415–425. https://doi.org/10.1007/s11033-016-3976-0
Yong L, He Y, Jin A et al (2014) Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc Natl Acad Sci U S A 111:E2414–E2422. https://doi.org/10.1073/pnas.1315605111
Zhuliang Y, Deng J, Li D et al (2020) Analysis of population structure and differentially selected regions in Guangxi native breeds by restriction site associated with DNA sequencing. G3: Genes, Genomes, Genetics 10:379–386. https://doi.org/10.1534/g3.119.400827
Zhang H, du ZQ, Dong JQ, Wang HX, Shi HY, Wang N, Wang SZ, Li H (2014) Detection of genome-wide copy number variations in two chicken lines divergently selected for abdominal fat content. BMC Genomics 15:517. https://doi.org/10.1186/1471-2164-15-517
Zhang H, Wang SZ, Wang ZP, da Y, Wang N, Hu XX, Zhang YD, Wang YX, Leng L, Tang ZQ, Li H (2012) A genome-wide scan of selective sweeps in two broiler chicken lines divergently selected for abdominal fat content. BMC Genomics 13:704. https://doi.org/10.1186/1471-2164-13-704
Zhang M, Ma X, Zhai Y, Zhang D, Sui L, Li W, Jiang R, Han R, Li G, Li Z, Wang Y, Tian Y, Kang X, Sun GR (2020) Comprehensive transcriptome analysis of lncRNAs reveals the role of lncAD in chicken intramuscular and abdominal adipogenesis. J Agric Food Chem 68:3678–3688. https://doi.org/10.1021/acs.jafc.9b07405
Zhao X, Mo D, Li A, Gong W, Xiao S, Zhang Y, Qin L, Niu Y, Guo Y, Liu X, Cong P, He Z, Wang C, Li J, Chen Y (2011) Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness. PLoS One 6:e19774. https://doi.org/10.1371/journal.pone.0019774
Funding
This work was supported in part by the National Natural Science Foundation of China (31660635) and the Science and Technology Major Project of Guangxi (GK AA17204027).
Author information
Authors and Affiliations
Contributions
Data curation: Cong Xiao, Tiantian Sun, Zuliang Yang, and Wenwen Xu. Formal analysis: Cong Xiao, Tiantian Sun, Zuliang Yang, and Xiurong Yang. Funding acquisition: Xiurong Yang. Investigation: Cong Xiao, Tiantian Sun, and Linghu Zeng. Methodology: Cong Xiao and Xiurong Yang. Project administration: Xiurong Yang. Resources: Jixian Deng and Xiurong Yang. Supervision: Jixian Deng and Xiurong Yang. Validation: Tiantian Sun and Juan Wang. Writing—original draft: Cong Xiao and Tiantian Sun. Writing—review and editing: Zuliang Yang and Xiurong Yang.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Highlights
1. RNA-seq of Nandan-Yao chicken in five tissues: abdominal fat, back skin, liver, pectoral muscle, leg muscle
2. Differentially expressed genes function enrichment analysis
3. Protein-protein interaction analysis of differentially expressed genes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Table S1-S5 represent the statistic of differentially expressed genes in the liver, back skin, pectoral muscle, leg muscle, abdominal fat, respectively. Table S6-S10 represents the statistics of GO enrichment items in the liver, back skin, pectoral muscle, leg muscle, abdominal fat, respectively. Table S11-S15 represents the statistics of KEGG enrichment items in the liver, back skin, pectoral muscle, leg muscle, abdominal fat, respectively.
ESM 1
(CSV 77 kb)
ESM 2
(CSV 91 kb)
ESM 3
(CSV 60 kb)
ESM 4
(CSV 73 kb)
ESM 5
(CSV 109 kb)
ESM 6
(CSV 242 kb)
ESM 7
(CSV 221 kb)
ESM 8
(CSV 202 kb)
ESM 9
(CSV 224 kb)
ESM 10
(CSV 294 kb)
ESM 11
(CSV 15 kb)
ESM 12
(CSV 16 kb)
ESM 13
(CSV 15 kb)
ESM 14
(CSV 14 kb)
ESM 15
(CSV 18 kb)
Rights and permissions
About this article
Cite this article
Xiao, C., Sun, T., Yang, Z. et al. Transcriptome landscapes of differentially expressed genes related to fat deposits in Nandan-Yao chicken. Funct Integr Genomics 21, 113–124 (2021). https://doi.org/10.1007/s10142-020-00764-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-020-00764-7