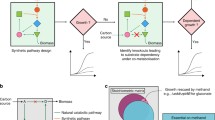
Abstract
Recent attempts to create synthetic Escherichia coli methylotrophs identified that de novo biosynthesis of amino acids, in the presence of methanol, presents significant challenges in achieving autonomous methylotrophic growth. Previously engineered methanol-dependent strains required co-utilization of stoichiometric amounts of co-substrates and methanol. As such, these strains could not be evolved to grow on methanol alone. In this work, we have explored an alternative approach to enable biosynthesis of all amino acids from methanol-derived carbon in minimal media without stoichiometric coupling. First, we identified that biosynthesis of threonine was limiting the growth of our methylotrophic E. coli. To address this, we performed adaptive laboratory evolution to generate a strain that grew efficiently in minimal medium with methanol and threonine. Methanol assimilation and growth of the evolved strain were analyzed, and, interestingly, we found that the evolved strain synthesized all amino acids, including threonine, from methanol-derived carbon. The evolved strain was then further engineered through overexpression of an optimized threonine biosynthetic pathway. We show that the resulting methylotrophic E. coli strain has a methanol-dependent growth phenotype with homoserine as co-substrate. In contrast to previous methanol-dependent strains, co-utilization of homoserine is not stoichiometrically linked to methanol assimilation. As such, future engineering of this strain and successive adaptive evolution could enable autonomous growth on methanol as the sole carbon source.
Key points
• Adaptive evolution of E. coli enables biosynthesis of all amino acids from methanol.
• Overexpression of threonine biosynthesis pathway improves methanol assimilation.
• Methanol-dependent growth is seen in minimal media with homoserine as co-substrate.




Similar content being viewed by others
Data availability
All strains generated in this study will be made available upon request.
References
Antoniewicz MR (2019) Synthetic methylotrophy: strategies to assimilate methanol for growth and chemicals production. Curr Opin Biotechnol 59:165–174. https://doi.org/10.1016/j.copbio.2019.07.001
Antonovsky N, Gleizer S, Noor E, Zohar Y, Herz E, Barenholz U, Zelcbuch L, Amram S, Wides A, Tepper N, Davidi D, Bar-On Y, Bareia T, Wernick DG, Shani I, Malitsky S, Jona G, Bar-Even A, Milo R (2016) Sugar synthesis from CO2 in Escherichia coli. Cell 166:115–125. https://doi.org/10.1016/j.cell.2016.05.064
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2. https://doi.org/10.1038/msb4100050
Bennett RK, Gonzalez JE, Whitaker WB, Antoniewicz MR, Papoutsakis ET (2018a) Expression of heterologous non-oxidative pentose phosphate pathway from Bacillus methanolicus and phosphoglucose isomerase deletion improves methanol assimilation and metabolite production by a synthetic Escherichia coli methylotroph. Metab Eng 45:75–85. https://doi.org/10.1016/j.ymben.2017.11.016
Bennett RK, Steinberg LM, Chen W, Papoutsakis ET (2018b) Engineering the bioconversion of methane and methanol to fuels and chemicals in native and synthetic methylotrophs. Curr Opin Biotechnol 50:81–93. https://doi.org/10.1016/j.copbio.2017.11.010
Bennett RK, Agee A, Har JRG, von Hagel B, Siu K-H, Antoniewicz MR, Papoutsakis ET (2020a) Triggering the stringent response enhances synthetic methanol utilization in Escherichia coli. Metab Eng 61:1–10. https://doi.org/10.1016/j.ymben.2020.04.007
Bennett RK, Dillon M, Har JRG, Agee A, von Hagel B, Rohlhill J, Antoniewicz MR, Papoutsakis ET (2020b) Engineering Escherichia coli for methanol-dependent growth on glucose for metabolite production. Metab Eng 60:45–55. https://doi.org/10.1016/j.ymben.2020.03.003
Chen CT, Chen FYH, Bogorad IW, Wu TY, Zhang R, Lee AS, Liao JC (2018) Synthetic methanol auxotrophy of Escherichia coli for methanol-dependent growth and production. Metab Eng 49:257–266. https://doi.org/10.1016/j.ymben.2018.08.010
Cotton CA, Claassens NJ, Benito-Vaquerizo S, Bar-Even A (2020) Renewable methanol and formate as microbial feedstocks. Curr Opin Biotechnol 62:168–180. https://doi.org/10.1016/j.copbio.2019.10.002
Diaz CAC, Bennett RK, Papoutsakis ET, Antoniewicz MR (2019) Deletion of four genes in Escherichia coli enables preferential consumption of xylose and secretion of glucose. Metab Eng 52:168–177. https://doi.org/10.1016/j.ymben.2018.12.003
Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. https://doi.org/10.1038/nmeth.1318
Gonzalez JE, Antoniewicz MR (2017) Tracing metabolism from lignocellulosic biomass and gaseous substrates to products with stable-isotopes. Curr Opin Biotechnol 43:86–95. https://doi.org/10.1016/j.copbio.2016.10.002
Gonzalez JE, Long CP, Antoniewicz MR (2017) Comprehensive analysis of glucose and xylose metabolism in Escherichia coli under aerobic and anaerobic conditions by 13C metabolic flux analysis. Metab Eng 39:9–18. https://doi.org/10.1016/j.ymben.2016.11.003
Gonzalez JE, Bennett RK, Papoutsakis ET, Antoniewicz MR (2018) Methanol assimilation in Escherichia coli is improved by co-utilization of threonine and deletion of leucine-responsive regulatory protein. Metab Eng 45:67–74. https://doi.org/10.1016/j.ymben.2017.11.015
Kim S, Lindner SN, Aslan S, Yishai O, Wenk S, Schann K, Bar-Even A (2020) Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol. https://doi.org/10.1038/s41589-020-0473-5
Krause SMB, Johnson T, Karunaratne YS, Fu Y, Beck DAC, Chistoserdova L, Lidstrom ME (2017) Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc Natl Acad Sci U S A 114:358–363. https://doi.org/10.1073/pnas.1619871114
Lee J, Lee B, Kim H (2003) Global analyses of transcriptomes and proteomes of a parent strain and an L-threonine-overproducing mutant strain. Microbiology 185:5442–5451. https://doi.org/10.1128/JB.185.18.5442
Long CP, Antoniewicz MR (2018) How adaptive evolution reshapes metabolism to improve fitness: recent advances and future outlook. Curr Opin Chem Eng 22:209–215. https://doi.org/10.1016/j.coche.2018.11.001
Long CP, Antoniewicz MR (2019) High-resolution 13C metabolic flux analysis. Nat Protoc 14:2856–2877. https://doi.org/10.1038/s41596-019-0204-0
Meyer F, Keller P, Hartl J, Gröninger OG, Kiefer P, Vorholt JA (2018) Methanol-essential growth of Escherichia coli. Nat Commun 9:1508. https://doi.org/10.1038/s41467-018-03937-y
Müller JEN, Heggeset TMB, Wendisch VF, Vorholt JA, Brautaset T (2015a) Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol. Appl Microbiol Biotechnol 99:535–551. https://doi.org/10.1007/s00253-014-6224-3
Müller JEN, Meyer F, Litsanov B, Kiefer P, Potthoff E, Heux S, Quax WJ, Wendisch VF, Brautaset T, Portais JC, Vorholt JA (2015b) Engineering Escherichia coli for methanol conversion. Metab Eng 28:190–201. https://doi.org/10.1016/j.ymben.2014.12.008
Rohlhill J, Sandoval NR, Papoutsakis ET (2017) Sort-Seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol. ACS Synth Biol 6:1584–1595. https://doi.org/10.1021/acssynbio.7b00114
Rohlhill J, Har JRG, Antoniewicz MR, Papoutsakis ET (2020) Improving synthetic methylotrophy via dynamic formaldehyde regulation of pentose phosphate pathway genes and redox perturbation. Metab Eng 57:247–255. https://doi.org/10.1016/j.ymben.2019.12.006
Sander T, Farke N, Diehl C, Kuntz M, Glatter T, Link H (2019) Allosteric feedback inhibition enables robust amino acid biosynthesis in E. coli by enforcing enzyme overabundance. Cell Syst 8:66-75.e8 . https://doi.org/10.1016/j.cels.2018.12.005
Whitaker WB, Sandoval NR, Bennett RK, Fast AG, Papoutsakis ET (2015) Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization. Curr Opin Biotechnol 33:165–175. https://doi.org/10.1016/j.copbio.2015.01.007
Whitaker WB, Jones JA, Bennett RK, Gonzalez JE, Vernacchio VR, Collins SM, Palmer MA, Schmidt S, Antoniewicz MR, Koffas MA, Papoutsakis ET (2017) Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli. Metab Eng 39:49–59. https://doi.org/10.1016/j.ymben.2016.10.015
Xu P, Vansiri A, Bhan N, Koffas MAG (2012) EPathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth Biol 1:256–266. https://doi.org/10.1021/sb300016b
Acknowledgments
The authors would like to thank Dr. Hannes Link for kindly providing the dysregulated E. coli strain, thrA*, from which the pThr* plasmid was constructed from, used in this study.
Funding
This work was supported by the Advanced Research Projects Agency-Energy (ARPA-E) Reducing Emissions using Methanotrophic Organisms for Transportation Energy (REMOTE) program (DE-AR0000432).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. JRGH and AA conducted the experiments. JRGH analyzed the data and wrote the manuscript. RKB, ETP, and MRA revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary materials associated with this article can be found in the online version.
ESM 1
(PDF 378 kb)
Rights and permissions
About this article
Cite this article
Har, J.R.G., Agee, A., Bennett, R.K. et al. Adaptive laboratory evolution of methylotrophic Escherichia coli enables synthesis of all amino acids from methanol-derived carbon. Appl Microbiol Biotechnol 105, 869–876 (2021). https://doi.org/10.1007/s00253-020-11058-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-11058-0