Abstract
We analyzed the prevalence and genetic characteristics of the extended-spectrum β-lactamases (ESBLs)-producing Enterobacterales isolated from adult patients hospitalized in the oncological center in 2019. Out of 9372 patients admitted to the hospital, 1373 had been in various medical facilities during the last year, which was an indication to perform a screening test for ESBL-producing Enterobacterales colonizing their gastrointestinal tract. In eighty-three patients (6.1%), 85 ESBL producers were detected. These isolates included the following: Escherichia coli (n = 67; 78.8%), Klebsiella pneumoniae (n = 14; 16.5%), Enterobacter cloacae cplx (n = 3; 3.5%), and Klebsiella oxytoca (n = 1; 1.2%). CTX-M-1-like enzymes were the most common ESBLs (n = 67; 78.8%). Two K. pneumoniae isolates (2/85; 2.4%) additionally produced New Delhi-metallo-β-lactamases (NDM). All isolates, except for K. oxytoca, were typed by pulsed-field gel electrophoresis (PFGE) and demonstrated high genetic diversity. The most prevalent phylogroups of E. coli were B2 group (n = 30; 44.8%), followed by A group (n = 25; 37.3%). These observations have motivated us to investigate the link between ESBL-E colonization and infection among patients with solid tumors.

Similar content being viewed by others
References
Gniadkowski M (2001) Evolution and epidemiology of extended-spectrum beta-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect 7:597–608
van den Bunt G, van Pelt W, Hidalgo L, Scharringa J, de Greeff SC et al (2019) Prevalence, risk factors and genetic characterisation of extended-spectrum beta-lactamase and carbapenemase-producing Enterobacteriaceae (ESBL-E and CPE): a community-based cross-sectional study, the Netherlands, 2014 to 2016. Euro Surveill. https://doi.org/10.2807/1560-7917.ES.2019.24.41.1800594
Xercavins M, Jiménez E, Padilla E, Riera M, Freixas N et al (2020) High clonal diversity of ESBL-producing Klebsiella pneumoniae isolates from clinical samples in a non-outbreak situation. Antimicrob Resist Infect Control, A cohort study. https://doi.org/10.1186/s13756-019-0661-9
Denkel LA, Maechler F, Schwab F, Kola A, Weber A et al (2019) Infections caused by extended-spectrum β-lactamase-producing Enterobacterales after rectal colonization with ESBL-producing Escherichia coli or Klebsiella pneumoniae. Clin Microbiol Infect. https://doi.org/10.1016/j.cmi.2019.11.025
Denis B, Lafaurie M, Donay JL, Fontaine JP, Oksenhendler E et al (2015) Prevalence, risk factors, and impact on clinical outcome of extended-spectrum beta-lactamase-producing Escherichia coli bacteraemia: a five-year study. Int J Inf Dis 39:1–6. https://doi.org/10.1016/j.ijid.2015.07.010
Karaiskos I, Giamarellou H (2020) Carbapenem-sparing strategies for ESBL producers: when and how. Antibiotics 9:61. https://doi.org/10.3390/antibiotics9020061
Kang ChI, Wi YM, Lee MY, Ko KS, Chung DR et al (2012) Epidemiology and risk factors of community onset infections caused by extended-spectrum ß-lactamase-producing Escherichia coli strains. J Clin Microbiol 50:312–317
Kern WV, Klose K, Jellen-Ritter AS, Oethinger M, Bohnert J et al (2005) Fluoroquinolone resistance of Escherichia coli at a cancer center: epidemiologic evolution and effects of discontinuing prophylactic fluoroquinolone use in neutropenic patients with leukemia. Eur J Clin Microbiol Infect Dis 24:111–118
Trecarichi EM, Giuliano G, Cattaneo C, Ballanti S, Criscuolo M et al (2019) Bloodstream infections caused by Escherichia coli in onco-haematological patients: risk factors and mortality in an Italian prospective survey. Haematologic malignancies associated bloodstream infections surveillance (HEMABIS) registry–Sorveglianza Epidemiologica Infezioni Fungine in Emopatie Maligne (SEIFEM) group, Italy. PLoS ONE 14(10):e0224465. https://doi.org/10.1371/journal.pone.0224465
Prevel R, Boyer A, M’Zali F, Lasheras A, Zahar JR et al (2019) Is systematic fecal carriage screening of extended-spectrum beta-lactamase-producing Enterobacteriaceae still useful in intensive care unit: a systematic review. Crit Care 23(1):170. https://doi.org/10.1186/s13054-019-2460-3
Carlet J (2012) The gut is the epicentre of antibiotic resistance. Antimicrob Resist Infect Control 1:39. https://doi.org/10.1186/2047-2994-1-39
Gupta A, Della-Latta P, Todd B, San Gabriel P, Haas J et al (2004) Outbreak of extended-spectrum ß-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit linked to artificial nails. Infect Control Hosp Epidmiol 25:210–215
European Committee on antimicrobial susceptibility testing. EUCAST guideline for the detection of resistance mechanisms and specific resistance of clinical and/or epidemiological importance. v 2.0 (2017-07-11). https://www.eucast.org/resistance_mechanisms/
Żabicka D, Baraniak A, Literacka E, Gniadkowski M, Hryniewicz (2017) Krajowy Ośrodek Referencyjny ds. Lekowrażliwości Drobnoustrojów: Wykrywanie karbapenemaz – zalecenia 2017. NIL, Warszawa
Empel J, Baraniak A, Literacka E, Mrowka A, Fiett J et al (2008) Molecular survey of β-lactamases conferring resistance to newer β-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob Agents Chemother 52:2449–2454
Empel J, Hrabak J, Kozinska A, Bergerova T, Urbaskova P et al (2010) DHA-1-producing Klebsiella pneumoniae in a teaching hospital in the Czech Republic. Microb Drug Resist 16:291–295
Woodford N, Fagan EJ, Ellington MJ (2006) Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother 57:154–155
Clermont O, Bonacorsi S, Bingen E (2000) Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. https://doi.org/10.1128/AEM.66.10.4555-4558.2000
Struelens MJ, Rost F, Deplano A, Maas A, Schwam V et al (1993) Pseudomonas aeruginosa and Enterobacteriaceae bacteremia after biliary endoscopy: an outbreak investigation using DNA macrorestriction analysis. Am J Med 95:489–498
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE et al (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239
Harris AD, Kotetishvili M, Shurland S, Johnson JA, Morris JG et al (2007) How important is patient-to-patient transmission in extended-spectrum beta-lactamase Escherichia coli acquisition. Am J Infect Control 35(2):97–101
Prevel R, Boyer A, M’Zali F, Cockenpot T, Lasheras A et al (2019) Extended spectrum beta-lactamase producing Enterobacterales faecal carriage in a medical intensive care unit: low rates of cross-transmission and infection. Antimicrob Resist Infect Control 8:112. https://doi.org/10.1186/s13756-019-0572-9
Aires-de-Sousa M, Lopes E, Gonçalves ML, Pereira AL, Machado E et al (2020) Intestinal carriage of extended-spectrum beta-lactamase producing Enterobacteriaceae at admission in a Portuquese hospital. Eur J Clin Microbiol Infect Dis 39(4):783–790
Arhoune B, Oumokhtar B, Hmami F, Barguigua A, Timinouni M et al (2017) Rectal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae among hospitalised neonates in a neonatal intensive care unit in Fez, Morocco. J Glob Antimicrob Resist 8:90–96. https://doi.org/10.1016/j.jgar.2016.11.004
Bozcal E, Eldem V, Aydemir S, Skurnik M (2018) The relationship between phylogenetic classification, virulence and antibiotic resistance of extraintestinal pathogenic Escherichia coli in Izmir province, Turkey. Peer J 6:e5470. https://doi.org/10.7717/peerj.5470
Iranpour D, Hassanpour M, Ansari H, Tajbakhsh S, Khamisipour G et al (2015) Phylogenetic groups of Escherichia coli strains from patients with urinary tract infection in Iran based on the new clermont phylotyping method. Biomed Res Int 2015:846219. https://doi.org/10.1155/2015/846219
Smati M, Clermont O, Gal FL, Schichmanoff O, Jauréguy F et al (2013) Real- time PCR for quantative analysis of human commensal Escherichia coli populations reveals a high frequency of sub-dominant phylogroups. Appl Environ Microbiol 79:5005–5012
Johnson JR, Delavari P, Kuskowski M, Stell AL (2001) Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 183:78–88
Lecointre G, Rachdi L, Darlu P, Denamur E (1998) Escherichia coli molecular phylogeny using the incongruence length difference test. Mol Biol Evol 15:1685–1695
Moreno E, Prats G, Sabaté M, Pérez T, Johnson JR et al (2006) Quinolone, fluoroquinolone and trimethoprim/sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. J Antimicrob Chemother 57(2):204–211
Farajzadah Sheikh A, Goodarzi H, Yadyad MJ, Aslani S, Amin M et al (2019) Virulence-associated genes and drug susceptibility patterns of uropathogenic Escherichia coli isolated from patients with urinary tract infection. Infect Drug Resist 12:2039–2047. https://doi.org/10.2147/IDR.S199764
Pitout JD, Laupland KB, Church DL, Menard ML, Johnson JR (2005) Virulence factors of Escherichia coli isolates that produce CTX-M-type extended-spectrum beta-lactamases. Antimicrob Agents Chemother 49(11):4667–4670
Chakraborty A, Saralaya V, Adhikari P, Shenoy S, Baliga S et al (2015) Characterization of Escherichia coli phylogenetic groups associated with extraintestinal infections in South Indian population. Ann Med Health Sci Res 5(4):241–246. https://doi.org/10.4103/2141-9248.160192
Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N et al (1999) The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67(2):546–553
Franiczek R, Sobieszczańska B, Turniak M, Kasprzykowska U, Krzyzanowska B et al (2012) ESBL-producing Escherichia coli isolated from children with acute diarrhea—antimicrobial susceptibility, adherence patterns and phylogenetic background. Adv Clin Exp Med 21(2):187–192
Chakraborty A, Adhikari P, Shenoy S, Saralaya V (2015) Genomic analysis and clinical importance of Escherichia coli isolate from patients with sepsis. Indian J Pathol Microbiol 58(1):22–26. https://doi.org/10.4103/0377-4929.151161
Rossolini GM, D’Andrea MM, Mugnaioli C (2008) The spread of CTX-M-type extended-spectrum ß-lactamases. Clin Microbiol Infect 14(Suppl. 1):33–41
Baraniak A, Fiett J, Hryniewicz W, Nordmann P, Gniadkowski M (2002) Ceftazidime-hydrolysing CTX-M-15 extended-spectrum β-lactamase (ESBL) in Poland. J Antimicrob Chemother 50(3):393–396. https://doi.org/10.1093/jac/dkf151
Pesesky MW, Hussain T, Wallace M, Wang B, Andleeb S et al (2015) KPC and NDM-1 genes in related Enterobacteriaceae strains and plasmids from Pakistan and the United States. Emerg Infect Dis 21(6):1034–1037. https://doi.org/10.3201/eid2106.141504
Baraniak A, Izdebski R, Fiett J, Gawryszewska I, Bojarska K et al (2016) NDM-producing enterobacteriaceae in Poland, 2012-14: inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. Antimicrob Chemother 71:85–91. https://doi.org/10.1093/jac/dkv282
Funding
This study had no funding.
Author information
Authors and Affiliations
Contributions
MS—conception, design, data analysis and interpretation, and wrote the manuscript, TN—acquisition and interpretation of clinical data, and ES and AB—performed the genetic characteristics of ESBL-E. Baraniak also critically reviewed the manuscript and revising for important intellectual content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest.
Ethics approval
The study received approval from the Bioethical Commission at the Collegium Medicum of Nicolaus Copernicus University in Toruń (KB approval number 216/2020).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2020_2334_MOESM1_ESM.tif
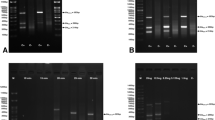
Fig 1 a-c Pulsed-field gel electrophoresis patterns (PFGE) of 67 isolates of E. coli. Identical isolates are marked with asterisks and dots. The remaining isolates represent unique PFGE patterns. M: λ ladder DNA (48.5 - 1,018 kb) (New England Biolabs, USA). (TIFF 2294 kb)
284_2020_2334_MOESM4_ESM.tif
PFGE gel image of 14 K. pneumoniae isolates. The asterisks denote indistinguishable isolates with NDM carbapenemases. Three isolates, slightly different from each other are marked with dots. M: λ ladder DNA (48.5 - 1,018 kb) (New England Biolabs, USA). (TIFF 764 kb)
284_2020_2334_MOESM5_ESM.tif
Fig 3 Three isolates of E. cloacae cplx with unique PFGE patterns. M: λ ladder DNA (48.5 - 1,018 kb) (New England Biolabs, USA). (TIFF 646 kb)
Rights and permissions
About this article
Cite this article
Szymankiewicz, M., Nowikiewicz, T., Stefaniuk, E. et al. Characteristics of ESBL-Producing Enterobacterales Colonizing the Gastrointestinal Tract in Patients Admitted to the Oncological Hospital. Curr Microbiol 78, 642–648 (2021). https://doi.org/10.1007/s00284-020-02334-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02334-3