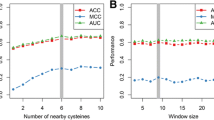
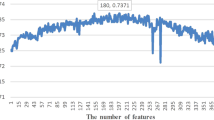
Abstract
Redox-sensitive cysteine (RSC) thiol contributes to many biological processes. The identification of RSC plays an important role in clarifying some mechanisms of redox-sensitive factors; nonetheless, experimental investigation of RSCs is expensive and time-consuming. The computational approaches that quickly and accurately identify candidate RSCs using the sequence information are urgently needed. Herein, an improved and robust computational predictor named IRC-Fuse was developed to identify the RSC by fusing of multiple feature representations. To enhance the performance of our model, we integrated the probability scores evaluated by the random forest models implementing different encoding schemes. Cross-validation results exhibited that the IRC-Fuse achieved accuracy and AUC of 0.741 and 0.807, respectively. The IRC-Fuse outperformed exiting methods with improvement of 10% and 13% on accuracy and MCC, respectively, over independent test data. Comparative analysis suggested that the IRC-Fuse was more effective and promising than the existing predictors. For the convenience of experimental scientists, the IRC-Fuse online web server was implemented and publicly accessible at http://kurata14.bio.kyutech.ac.jp/IRC-Fuse/.




Similar content being viewed by others
References
Chinenov Y, Schmidt T, Yang XY, Martin ME (1998) Identification of redox-sensitive cysteines in GA-binding protein-alpha that regulate DNA binding and heterodimerization. The Journal of biological chemistry 273(11):6203–6209
Anderson LE, Li D, Prakash N, Stevens FJ (1995) Identification of potential redox-sensitive cysteines in cytosolic forms of fructosebisphosphatase and glyceraldehyde-3-phosphate dehydrogenase. Planta 196(1):118–124
Wouters MA, Fan SW, Haworth NL (2010) Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid Redox Signal 12(1):53–91
Herrmann JM, Becker K, Dick TP (2015) Highlight: dynamics of thiol-based redox switches. Biol Chem 396(5):385–387
Antelmann H, Helmann JD (2011) Thiol-based redox switches and gene regulation. Antioxid Redox Signal 14(6):1049–1063
Brandes N, Schmitt S, Jakob U (2009) Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal 11(5):997–1014
Sun MA, Zhang Q, Wang Y, Ge W, Guo D (2016) Prediction of redox-sensitive cysteines using sequential distance and other sequence-based features. BMC Bioinformatics 17(1):316
Longen S, Beck KF, Pfeilschifter J (2016) H2S-induced thiol-based redox switches: Biochemistry and functional relevance for inflammatory diseases. Pharmacol Res 111:642–651
Groitl B, Jakob U (2014) Thiol-based redox switches. Biochem Biophys Acta 1844(8):1335–1343
Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG, Szypowska A, Meppelink A, Brenkman AB, Yodoi J et al (2009) Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol 5(9):664–672
Sanchez R, Riddle M, Woo J, Momand J (2008) Prediction of reversibly oxidized protein cysteine thiols using protein structure properties. Protein science : a publication of the Protein Society 17(3):473–481
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Charoenkwan P, Nantasenamat C, Hasan MM, Shoombuatong W (2020) iTTCA-Hybrid: Improved and robust identification of tumor T cell antigens by utilizing hybrid feature representation. Anal Biochem 599:113747
Hasan MM, Rashid MM, Khatun MS, Kurata H (2019) Computational identification of microbial phosphorylation sites by the enhanced characteristics of sequence information. Sci Rep 9(1):8258
Hasan MM, Manavalan B, Khatun MS, Kurata H (2019) Prediction of S-nitrosylation sites by integrating support vector machines and random forest. Mol Omics 15(6):451–458
Hasan MM, Khatun MS, Mollah MNH, Yong C, Guo D (2017) A systematic identification of species-specific protein succinylation sites using joint element features information. Int J Nanomedicine 12:6303–6315
Chen K, Jiang Y, Du L, Kurgan L (2009) Prediction of integral membrane protein type by collocated hydrophobic amino acid pairs. J Comput Chem 30(1):163–172
Khatun MS, Hasan MM, Kurata H (2019) PreAIP: Computational Prediction of Anti-inflammatory Peptides by Integrating Multiple Complementary Features. Front Genet 10:129
Hasan MM, Kurata H (2018) GPSuc: Global Prediction of Generic and Species-specific Succinylation Sites by aggregating multiple sequence features. PLoS ONE 13(10):e0200283
Hasan MM, Khatun MS, Mollah MNH, Yong C, Dianjing G (2018) NTyroSite: Computational Identification of Protein Nitrotyrosine Sites Using Sequence Evolutionary Features. Molecules. https://doi.org/10.3390/molecules23071667
Hasan MM, Zhou Y, Lu X, Li J, Song J, Zhang Z (2015) Computational Identification of Protein Pupylation Sites by Using Profile-Based Composition of k-Spaced Amino Acid Pairs. PLoS ONE 10(6):e0129635
Chen W, Tang H, Ye J, Lin H, Chou KC (2016) iRNA-PseU: Identifying RNA pseudouridine sites. Molecular therapy Nucleic acids 5:e332
Liu B, Liu F, Wang X, Chen J, Fang L, Chou KC (2015) Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucleic Acids Res 43(W1):W65-71
Adilina S, Farid DM, Shatabda S (2019) Effective DNA binding protein prediction by using key features via Chou’s general PseAAC. J Theor Biol 460:64–78
Charoenkwan P, Schaduangrat N, Nantasenamat C, Piacham T, Shoombuatong W (2019) iQSP: A Sequence-Based Tool for the Prediction and Analysis of Quorum Sensing Peptides via Chou’s 5-Steps Rule and Informative Physicochemical Properties. Int J Mol Sci. https://doi.org/10.3390/ijms21010075
Manavalan B, Hasan MM, Basith S, Gosu V, Shin TH, Lee G (2020) Empirical Comparison and Analysis of Web-Based DNA N4-Methylcytosine Site Prediction Tools. Molecular Therapy-Nucleic Acids 22:406–420
Maclin R, Opitz D (1999) Popular ensemble methods: An empirical study. Journal of Artificial Intelligence Research. https://doi.org/10.1613/jair.614
Polikar R (2006) Ensemble based systems in decision making. Circuits and systems magazine, IEEE 6(3):21–45
Rokach L (2010) Ensemble-based classifiers. Artif Intell Rev 33(1–2):1–39
Khatun S, Hasan M, Kurata H (2019) Efficient computational model for identification of antitubercular peptides by integrating amino acid patterns and properties. FEBS Lett. https://doi.org/10.1002/1873-3468.13536
Charoenkwan P, Kanthawong S, Schaduangrat N, Yana J, Shoombuatong W (2020) PVPred-SCM: Improved Prediction and Analysis of Phage Virion Proteins Using a Scoring Card Method. Cells. https://doi.org/10.3390/cells9020353
Charoenkwan P, Shoombuatong W, Lee HC, Chaijaruwanich J, Huang HL, Ho SY (2013) SCMCRYS: predicting protein crystallization using an ensemble scoring card method with estimating propensity scores of P-collocated amino acid pairs. PLoS ONE 8(9):e72368
Basith S, Manavalan B, Hwan Shin T, Lee G (2020) Machine intelligence in peptide therapeutics: A next-generation tool for rapid disease screening. Med Res Rev. https://doi.org/10.1002/med.21658
Manavalan B, Basith S, Shin TH, Wei L, Lee G (2019) mAHTPred: a sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics 35(16):2757–2765
Mosharaf MP, Hassan MM, Ahmed FF, Khatun MS, Moni MA, Mollah MNH (2020) Computational prediction of protein ubiquitination sites mapping on Arabidopsis thaliana. Comput Biol Chem 85:107238
Liaw A, Wiener M (2002) Classification and regression by randomForest. R news 2(3):18–22
Manavalan B, Shin TH, Lee G (2018) PVP-SVM: Sequence-Based Prediction of Phage Virion Proteins Using a Support Vector Machine. Front Microbiol 9:476
Manavalan B, Shin TH, Lee G (2018) DHSpred: support-vector-machine-based human DNase I hypersensitive sites prediction using the optimal features selected by random forest. Oncotarget 9(2):1944–1956
Alam MA, Komori O, Deng HW, Calhoun VD, Wang YP (2019) Robust kernel canonical correlation analysis to detect gene-gene co-associations: A case study in genetics. J Bioinform Comput Biol 17(4):1950028
Alam MA, Lin HY, Deng HW, Calhoun VD, Wang YP (2018) A kernel machine method for detecting higher order interactions in multimodal datasets: Application to schizophrenia. J Neurosci Methods 309:161–174
Alam MA, Fukumizu K, Wang YP (2018) Influence Function and Robust Variant of Kernel Canonical Correlation Analysis. Neurocomputing 304:12–29
Ahamad MM, Aktar S, Rashed-Al-Mahfuz M, Uddin S, Lio P, Xu H, Summers MA, Quinn JMW, Moni MA (2020) A machine learning model to identify early stage symptoms of SARS-Cov-2 infected patients. Expert Syst Appl 160:113661
Liaw A (2002) Wiener: Classification and regression by random forest. R news 2:18–22
Su R, Hu J, Zou Q, Manavalan B, Wei L (2019) Empirical comparison and analysis of web-based cell-penetrating peptide prediction tools. Brief Bioinform. https://doi.org/10.1093/bib/bby124
Shoombuatong W, Schaduangrat N, Pratiwi R, Nantasenamat C (2019) THPep: A machine learning-based approach for predicting tumor homing peptides. Comput Biol Chem 80:441–451
Schaduangrat N, Nantasenamat C, Prachayasittikul V, Shoombuatong W (2019) Meta-iAVP: A Sequence-Based Meta-Predictor for Improving the Prediction of Antiviral Peptides Using Effective Feature Representation. Int J Mol Sci. https://doi.org/10.3390/ijms20225743
Win TS, Malik AA, Prachayasittikul V (2017) JE SW, Nantasenamat C, Shoombuatong W: HemoPred: a web server for predicting the hemolytic activity of peptides. Future medicinal chemistry 9(3):275–291
Manavalan B, Shin TH, Kim MO, Lee G (2018) PIP-EL: A New Ensemble Learning Method for Improved Proinflammatory Peptide Predictions. Front Immunol 9:1783
Hasan MM, Khatun MS, Kurata H (2020) iLBE for Computational Identification of Linear B-cell Epitopes by Integrating Sequence and Evolutionary Features. Genomics Proteomics Bioinformatics. https://doi.org/10.1016/j.gpb.2019.04.004
Charoenkwan P, Yana J, Nantasenamat C, Hasan MM, Shoombuatong W (2020) iUmami-SCM: A Novel Sequence-Based Predictor for Prediction and Analysis of Umami Peptides Using a Scoring Card Method with Propensity Scores of Dipeptides. J Chem Inf Model. https://doi.org/10.1021/acs.jcim.0c00707
Charoenkwan P, Kanthawong S, Nantasenamat C, Hasan MM, Shoombuatong W (2020) iDPPIV-SCM: A Sequence-Based Predictor for Identifying and Analyzing Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides Using a Scoring Card Method. J Proteome Res 19(10):4125–4136
Vacic V, Iakoucheva LM, Radivojac P (2006) Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics 22(12):1536–1537
Charoenkwan P, Kanthawong S, Nantasenamat C, Hasan MM, Shoombuatong W (2020) iAMY-SCM: Improved prediction and analysis of amyloid proteins using a scoring card method with propensity scores of dipeptides. Genomics. https://doi.org/10.1016/j.ygeno.2020.09.065
Provost F: Machine Learning from Imbalanced Data Sets 101. AAAI Workshop on learning from imbalanced data set 2000:1–3.
Lin C-J, Chen Y-W (2003) Combining SVMs with Various Feature Selection Strategies. In: Lin C-J, Chen Y-W (eds) NIPS 2003 feature selection challenge. Springer, Berlin, pp 1–10
Shi SP, Qiu JD, Sun XY, Suo SB, Huang SY, Liang RP (2012) PLMLA: prediction of lysine methylation and lysine acetylation by combining multiple features. Mol BioSyst 8(5):1520–1527
Li Y, Wang M, Wang H, Tan H, Zhang Z, Webb GI, Song J (2014) Accurate in silico identification of species-specific acetylation sites by integrating protein sequence-derived and functional features. Scientific reports 4:5765
Hasan MM, Yang S, Zhou Y, Mollah MN (2016) SuccinSite: a computational tool for the prediction of protein succinylation sites by exploiting the amino acid patterns and properties. Mol Biosyst 12(3):786–795
Boopathi V, Subramaniyam S, Malik A, Lee G, Manavalan B, Yang DC (2019) mACPpred: A Support Vector Machine-Based Meta-Predictor for Identification of Anticancer Peptides. Int J Mol Sci. https://doi.org/10.3390/ijms20081964
Manavalan B, Basith S, Shin TH, Wei L, Lee G (2019) Meta-4mCpred: A Sequence-Based Meta-Predictor for Accurate DNA 4mC Site Prediction Using Effective Feature Representation. Mol Ther Nucleic Acids 16:733–744
Manavalan B, Basith S, Shin TH, Wei L, Lee G (2019) AtbPpred: A Robust Sequence-Based Prediction of Anti-Tubercular Peptides Using Extremely Randomized Trees. Comput Struct Biotechnol J 17:972–981
Hasan MM, Manavalan B, Khatun MS, Kurata H (2019) i4mC-ROSE, a bioinformatics tool for the identification of DNA N4-methylcytosine sites in the Rosaceae genome. Int J Biol Macromol. https://doi.org/10.1016/j.ijbiomac.2019.12.009
Hasan MM, Guo D, Kurata H (2017) Computational identification of protein S-sulfenylation sites by incorporating the multiple sequence features information. Mol Biosyst 13(12):2545–2550
Chen Z, Zhao P, Li F, Wang Y, Smith AI, Webb GI, Akutsu T, Baggag A, Bensmail H, Song J (2019) Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences. Brief Bioinform. https://doi.org/10.1093/bib/bbz112
Charoenkwan P, Anuwongcharoen N, Nantasenamat C, Hasan MM, Shoombuatong W (2020) In silico approaches for the prediction and analysis of antiviral peptides: a review. Curr Pharm Des. https://doi.org/10.2174/1381612826666201102105827
Manavalan B, Basith S, Shin TH, Lee G (2020) Computational prediction of species-specific yeast DNA replication origin via iterative feature representation. Brief Bioinform. https://doi.org/10.1093/bib/bbaa304
Chowdhury SY, Shatabda S, Dehzangi A (2017) iDNAProt-ES: Identification of DNA-binding Proteins Using Evolutionary and Structural Features. Scientific reports 7(1):14938
Khatun MS, Hasan MM, Shoombuatong W, Kurata H (2020) ProIn-Fuse: improved and robust prediction of proinflammatory peptides by fusing of multiple feature representations. J Comput Aided Mol Des. https://doi.org/10.1007/s10822-020-00343-9
Hasan MM, Schaduangrat N, Basith S, Lee G, Shoombuatong W, Manavalan B (2020) HLPpred-Fuse: improved and robust prediction of hemolytic peptide and its activity by fusing multiple feature representation. Bioinformatics 36(11):3350–3356
Hasan MM, Basith S, Khatun MS, Lee G, Manavalan B, Kurata H (2020) Meta-i6mA: an interspecies predictor for identifying DNA N6-methyladenine sites of plant genomes by exploiting informative features in an integrative machine-learning framework. Brief Bioinform. https://doi.org/10.1093/bib/bbaa202
Rahman MS, Aktar U, Jani MR, Shatabda S (2019) iPromoter-FSEn: Identification of bacterial sigma(70) promoter sequences using feature subspace based ensemble classifier. Genomics 111(5):1160–1166
Muhammod R, Ahmed S, Md Farid D, Shatabda S, Sharma A, Dehzangi A (2019) PyFeat: a Python-based effective feature generation tool for DNA. RNA and protein sequences Bioinformatics 35(19):3831–3833
Acknowledgements
This work is supported by the Grant-in-Aid for JSPS Research Fellow (19F19377) from Japan Society for the Promotion of Science (JSPS). Partially supported from Japan Society for the Promotion of Science by Grant-in-Aid for Scientific Research (B) (19H04208) and by the developing key technologies for discovering and manufacturing pharmaceuticals used for next-generation treatments and diagnoses both from the Ministry of Economy, Trade and Industry, Japan (METI) and from Japan Agency for Medical Research and Development (AMED).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hasan, M.M., Alam, M.A., Shoombuatong, W. et al. IRC-Fuse: improved and robust prediction of redox-sensitive cysteine by fusing of multiple feature representations. J Comput Aided Mol Des 35, 315–323 (2021). https://doi.org/10.1007/s10822-020-00368-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00368-0