Abstract
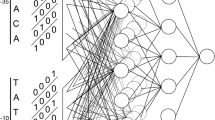
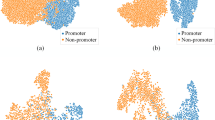
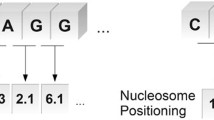
This paper is about a new computational strategy to find the promoter sequence which is a transcription starting factor in DNA without which the gene sequence cannot be activated for further protein synthesis process. Genes are the hereditary source of organisms’ functionality of living things. Sequence of activities is carried out to reveal the protein production process hidden in the genes. In this regard promoter prediction is essential to bring out correctness of protein synthesis in decoding the genetical information. Also it enables the biologists to find the genetical malfunctions. Nowadays numerous computational methods which have been proposed are helpful to the biologist in related tasks. Earlier works applied Gibb sampling method, Markov Chain Model and machine learning strategies to predict the promoters. In this paper a new computational strategy named as GSCNN is introduced which combines Gibb sampling with Convolution Neural Network (CNN). This model uses Gibb sampling method to select the best profile features of DNA and extract the scores as feature vector with the help of position frequency matrix. Those extracted feature vectors are further fed into Convolution Neural Network to predict the sigma (σ) 54 promoter of bacterial genome. The performance of the proposed system is evaluated with the help of Bacillus subtilis NC_000964 dataset. The sigma (σ) 54 promoter prediction using the proposed GSCNN method achieved significant improvement in performance metrics compared with the traditional machine learning algorithms such as Decision Tree (DT), K-Nearest Neighbor(KNN), Random Forest(RF) and Support Vector Machine(SVM).




Similar content being viewed by others
References
Shahmuradov IA, Razali RM, Bougouffa S, Radovanovic A, Bajic VB (2017) bTSSfinder: a novel tool for the prediction of promoters in cyanobacteria and Escherichia coli. Bioinformatics 33(3):334–340. https://doi.org/10.1093/bioinformatics/btw629
Lin H, Deng EZ, Ding H, Chen W, Chou KC (2014) iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res 42(21):12961–129672. https://doi.org/10.1093/nar/gku1019 ((ISBN: 0305-1048))
Barrios H, Valderrama B, Morett E (1999) Compilation and analysis of σ54 dependent promoter sequences. Nucleic Acids Res 27(22):4305–4313. https://doi.org/10.1093/nar/27.22.4305
Mallios RR, Ojcius DM, Ardel DH (2009) An iterative strategy combining biophysical criteria and duration hidden Markov models for structural predictions of Chlamydia trachomatis σ66 promoters. BMC Bioinform 10:27. https://doi.org/10.1186/1471-2105-10-271(ISBN:1471-2105)
Shida K (2006) GibbST: a Gibb sampling method for motif discovery with enhanced resistance to local optima. BMC Bioinform 7:486. https://doi.org/10.1186/1471-2105-7-486
Zhou T, Shen N, Yang L, Abe N, Horton J, Mann RS, Bussemaker HJ, Gordân R, Rohs R (2015) Quantitative modeling of transcription factor binding specificities using DNA shape. Proc Natl Acad Sci 112(15):4654–4659. https://doi.org/10.1073/pnas.1422023112 ((ISBN: 0027-8424))
Premalatha C, Aravindan C, Kannan K (2011) Promoter prediction in eukaryotes using soft computing techniques. IEEE Conf Recent Adv Intell Comput Syst. https://doi.org/10.1109/RAICS.2011.6069368ISBN:978-1-4244-9478-1
He W, Jia C, Duan Y, Zou Q (2017) 70ProPred: a predictor for discovering σ70 promoters based on combining multiple features. In: 11th International Conference on Systems Biology (ISB 2017) Shenzhen, China, https://doi.org/10.1007/978-3-319-68759-9 (ISBN: 978–3–319–68758–2 ISBN: 0302–9743)
Li Q-Z, Lin H (2006) The recognition and prediction of sigma70 promoters in Escherichia coli K-12. J Theor Biol 242:135–141. https://doi.org/10.1016/j.jtbi.2006.02.007ISBN:0022-5193
Gan Y, Guan J, Zhou S (2012) A comparison study on feature selection of DNA structural properties for promoter prediction. BMC Bioinform 13:4. https://doi.org/10.1186/1471-2105-13-4ISBN:1471-2105
Gusmao EG, Dieterich C, Zenke M, Costa IG (2014) Detection of active transcription factor binding sites with the combination of DNase hypersensitivity and histone modifications. Bioinformatics 30(22):3143–3151. https://doi.org/10.1093/bioinformatics/btu519 ((ISBN: 1367-4803))
Bland C, Newsome AS, Markovets AA (2010) Promoter prediction in E. coli based on SIDD profiles and artificial neural networks. In: 7th Annual MCBIOS Conference Bioinformatics: Systems, Biology, Informatics and Computation Jonesboro, AR, USA ; February 2010, https://doi.org/10.1186/1471-2105-11-S6-S17
Liu D, Xiang X, DasGupta B, Zhang H (2006) Motif Discoveries in Unaligned Molecular Sequences Using Self-Organizing Neural Networks. IEEE Trans Neural Netw. https://doi.org/10.1109/TNN.2006.875987 ((ISBN: 1045-9227))
Abbass MM, Bahig HM (2013) An efficient algorithm to identify DNA motifs. Math Compu 7:387–399. https://doi.org/10.1007/s11786-013-0165-6
Makolo AU, Lamidi UA (2018) Motif discovery in DNA sequences using improved gibbs (i Gibbs) sampling algorithm. J Comput Sci Syst Biol 11:5. https://doi.org/10.4172/jcsb.1000288
Frith MC, Hansen U, Spouge JL, Weng Z (2004) Finding functional sequence elements by multiple local alignment. Nuclic Acids Res 32(1):189–200. https://doi.org/10.1093/nar/gkh169
Kilpatrick AM, Ward B, Aitken S (2013) MCOIN: a novel heuristic for determining transcription factor binding site motif width. Algorithm Mol Biol 8:16 http://www.almob.org/content/8/1/16; https://doi.org/10.1186/1748-7188-8-16 (ISBN: 1748–7188)
Liu B, Han L, Liu X, Wu J, Ma Q (2018) Computational prediction of sigma-54 promoters in bacterial genomes by integrating motif finding and machine learning strategies. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2018.2816032 ((ISBN: 1545-5963))
Meng G, Mosig A, Vingron M (2010) A computational evaluation of over-representation of regulatory motifs in the promoter regions of differentially expressed genes. BMC Bioinform 11:267. https://doi.org/10.1186/1471-2105-11-267 ((ISBN: 1471-2105))
Lin H, Li Q-Z (2011) Eukaryotic and prokaryotic promoter prediction using hybrid approach. Theory Biosci 130:91–100. https://doi.org/10.1007/s12064-010-0114-8 ((ISBN: 1367-4803))
Di Salvo M, Pinatel E, Tala A, Fondi M, Peano C, Alifano P (2018) G4PromFinder: an algorithm for predicting transcription promoters in GC-rich bacterial genomes based on AT-rich elements and G-quadruplex motifs. BMC Bioinform 19:36. https://doi.org/10.1186/s12859-018-2049-xISBN:1471-2105
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sasikala, S., Ratha Jeyalakshmi, T. GSCNN: a composition of CNN and Gibb Sampling computational strategy for predicting promoter in bacterial genomes. Int. j. inf. tecnol. 13, 493–499 (2021). https://doi.org/10.1007/s41870-020-00565-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41870-020-00565-y