Abstract
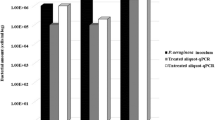
The arthroconidial yeasts Magnusiomyces capitatus and M. clavatus are emerging opportunistic pulmonary pathogens. They are closely related and difficult to distinguish based on morphological and physiological traits. We applied an SYBR® green-based quantitative PCR (qPCR) assay to identify the species. We analyzed 30 reference strains originating from clinical and environmental sources by targeting the Rpb2 gene encoding the second largest subunit of RNA polymerase II. The qPCR assays were tested by direct identification of M. capitatus and M. clavatus in spiked sputum and household dishwasher swabs, respectively, as models for clinical and environmental samples. The assays were proved to be reliable for species-level identification of both species, with 100% sensitivity and 100% specificity, lowest inter-assay deviations (RSDr ≤ 1.65%, R2 values >0.99), detection limit of 10 theoretical copy number of target DNA, and detection cell limit of ≥5000 yeast cells from spiked sputum samples. The developed qPCR assay is a practical molecular approach for the detection of M. capitatus and M. clavatus that can be used as a stand-alone assay or in conjunction with culture-dependent approaches.





Similar content being viewed by others
References
de Hoog GS, Smith MT. The ribosomal gene phylogeny and species delimitation in Geotrichum and its teleomorphs. Stud Mycol. 2004;50:489–515.
de Hoog GS, Smith MT, Guého E. A revision of the genus Geotrichum and its teleomorphs. Stud Mycol. 1986;29:1–131.
Kaplan E, Al-Hatmi AMS, Ilkit M, et al. Molecular diagnostics of arthroconidial yeasts, frequent pulmonary opportunists. J Clin Microbiol. 2018;56(1):e01427-e1517.
García-Ruiz JC, López-Soria L, Olazábal I, et al. Invasive infections caused by Saprochaete capitata in patients with haematological malignancies: report of five cases and review of the antifungal therapy. Rev Iberoam Micol. 2013;30(4):248–55.
Vaux S, Criscuolo A, Desnos-Ollivier M, et al. Multicenter outbreak of infections by Saprochaete clavata, an unrecognized opportunistic fungal pathogen. mBio. 2014;5:e02309-14.
Mazzocato S, Marchionni E, Fothergill AW, et al. Epidemiology and outcome of systemic infections due to Saprochaete capitata: case report and review of the literature. Infection. 2015;43(2):211–5.
Schuermans C, van Bergen M, Coorevits L, et al. Breakthrough Saprochaete capitata infections in patients receiving echinocandins: case report and review of the literature. Med Mycol. 2011;49(4):414–8.
Birrenbach T, Bertschy S, Aebersold F, et al. Emergence of Blastoschizomyces capitatus yeast infections, Central Europe. Emerg Infect Dis. 2012;18(1):98–101.
Zalar P, Novak M, de Hoog GS, Gunde-Cimerman N. Dishwashers: a man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biol. 2011;115(10):997–1007.
Döğen A, Kaplan E, Öksüz Z, Serin MS, Ilkit M, de Hoog GS. Dishwashers are a major source of human opportunistic yeast-like fungi in indoor environments in Mersin. Turkey Med Mycol. 2013;51(5):493–8.
Gümral R, Özhak-Baysan B, Tümgör A, et al. Dishwashers provide a selective extreme environment for human-opportunistic yeast-like fungi. Fungal Divers. 2016;76(1):1–9.
Zupančič J, Novak Babic M, Zalar P, Gunde-Cimerman N. The black yeast Exophiala dermatitidis and other selected opportunistic human fungal pathogens spread from dishwashers to kitchens. PLoS ONE. 2016;11(2):e0148166.
Tanuskova D, Horakova J, Svec P, et al. First case of invasive Magnusiomyces capitatus infection in Slovakia. Med Mycol Case Rep. 2017;16:12–5.
Leoni M, Riccardi N, Rotulo GA, et al. Magnusiomyces clavatus infection in a child after allogeneic hematotopoetic stem cell transplantation: diagnostic and therapeutic implications. Med Mycol Case Rep. 2018;23:65–7.
Buchta V, Bolehovská R, Hovorková E, Cornely OA, Seidel D, Žák P. Saprochaete clavata invasive infections: a new threat to hematological-oncological patients. Front Microbiol. 2019;10:2196.
Pamidimukkala U, Kancharla A, Sudhaharan S, et al. Isolation of the rare opportunistic yeast Saprochaete capitata from clinical samples-experience from a tertiary care hospital in southern India and a brief review of the literature. J Clin Diagn Res. 2017;11(9):36–42.
Esposto MC, Prigitano A, Lo Cascio G, et al. Yeast-like filamentous fungi: molecular identification and in vitro susceptibility study. Med Mycol. 2019;57(7):909–13.
Desnos-Ollivier M, Blanc C, Garcia-Hermoso D, Hoinard D, Alanio A, Dromer F. Misidentification of Saprochaete clavata as Magnusiomyces capitatus in clinical isolates: utility of internal transcribed spacer sequencing and matrix-assisted laser desorption ionization: time of flight mass spectrometry and importance of reliable databases. J Clin Microbiol. 2014;52(6):2196–8.
Koç AN, Atalay MA, Timur D, Demir G, Kaynar L. Molecular epidemiology and antifungal susceptibility of Saprochaete capitata (Blastoschizomyces capitatus) isolates causing nosocomial infection in Kayseri/Turkey. Infect Dis (Lond). 2016;48(8):596–603.
Springer J, McCormick Smith I, Hartmann S, et al. Identification of Aspergillus and Mucorales in formalin-fixed, paraffin-embedded tissue samples: comparison of specific and broad-range fungal qPCR assays. Med Mycol. 2019;57(3):308–13.
Salehi E, Hedayati MT, Zoll J, et al. Discrimination of aspergillosis, mucormycosis, fusariosis, and scedosporiosis in formalin-fixed paraffin-embedded tissue specimens by use of multiple real-time quantitative PCR assays. J Clin Microbiol. 2016;54(11):2798–803.
Krohn S, Zeller K, Böhm S, et al. Molecular quantification and differentiation of Candida species in biological specimens of patients with liver cirrhosis. PLoS ONE. 2018;13(6):e0197319.
Fidler G, Leiter E, Kocsube S, Biro S, Paholcsek M. Validation of a simplex PCR assay enabling reliable identification of clinically relevant Candida species. BMC Infect Dis. 2018;18(1):393.
Springer J, Lackner M, Ensinger C, et al. Clinical evaluation of a Mucorales-specific real-time PCR assay in tissue and serum samples. J Med Microbiol. 2016;65(12):1414–21.
Al-Hatmi AM, Bonifaz A, de Hoog GS, et al. Keratitis by Fusarium temperatum, a novel opportunist. BMC Infect Dis. 2014;14:588.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Owczarzy R, Tataurov AV, Wu Y, et al. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:163–9.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134.
Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22.
European Network of GMO Laboratories. Verification of analytical methods for GMO testing when implementing interlaboratory validated methods. Luxembourg: Joint Research Centre, Luxembourg; 2011.
Blakely T, Salmond C. Probabilistic record linkage and a method to calculate the positive predictive value. Int J Epidemiol. 2002;31(6):1246–52.
Brejová B, Lichancová H, Brázdovič F, et al. Genome sequence of the opportunistic human pathogen Magnusiomyces capitatus. Curr Genet. 2019;65(2):539–60.
Turin L, Riva F, Galbiati G, Cainelli T. Fast, simple and highly sensitive double-rounded polymerase chain reaction assay to detect medically relevant fungi in dermatological specimens. Eur J Clin Invest. 2000;30(6):511–8.
Everaerts S, Lagrou K, Vermeersch K, Dupont LJ, Vanaudenaerde BM, Janssens W. Aspergillus fumigatus detection and risk factors in patients with COPD-Bronchiectasis overlap. Int J Mol Sci. 2018;19(2):e523.
Liu CM, Kachur S, Dwan MG, et al. FungiQuant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 2012;12:255.
Kaplan E, Ilkit M, de Hoog GS. Comparison of the rolling circle amplification and ligase-dependent reaction methods for the identification of opportunistic Exophiala species. Med Mycol. 2018;56(6):759–69.
Prévost-Bouré NC, Christen R, Dequiedt S, et al. Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS ONE. 2011;6(9):e24166.
Fredricks DN, Smith C, Meier A. Comparison of six DNA extraction methods for recovery of fungal DNA as assessed by quantitative PCR. J Clin Microbiol. 2005;43(10):5122–8.
Dalla-Costa LM, Morello LG, Conte D, et al. Comparison of DNA extraction methods used to detect bacterial and yeast DNA from spiked whole blood by real-time PCR. J Microbiol Methods. 2017;140:61–6.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: EK, AD, SHP, MI, GSH. Performed the experiments: EK, DA, AD, SHP, RG. Analyzed the data: EK, DA, AD, SHP, RG, FH, MI, GSH. Contributed reagents/materials/analysis tools: EK, DA, AD, SHP, RG, FH, MI, GSH. Wrote the paper: EK, DA, AD, SHP, RG, FH, MI, GSH.
Corresponding authors
Ethics declarations
Conflict of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of this paper.
Ethics Statement
Ethical approval and patient consensus were not necessary because the study design was largely based on reference strains. Non-reference clinical isolates originated from anonymized clinical samples obtained during routine laboratory activity.
Additional information
Handling Editor: Vishnu Chaturvedi
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kaplan, E., Aktaş, D., Döğen, A. et al. Development and Analysis of qPCR for the Identification of Arthroconidial Yeasts of the Genus Magnusiomyces. Mycopathologia 186, 41–51 (2021). https://doi.org/10.1007/s11046-020-00510-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-020-00510-4