Abstract
Histopathological image contains rich pathological information that is valued for the aided diagnosis of many diseases such as cancer. An important issue in histopathological image classification is how to learn a high-quality discriminative dictionary due to diverse tissue pattern, a variety of texture, and different morphologies structure. In this paper, we propose a discriminative dictionary learning algorithm with pairwise local constraints (PLCDDL) for histopathological image classification. Inspired by the one-to-one mapping between dictionary atom and profile, we learn a pair of discriminative graph Laplacian matrices that are less sensitive to noise or outliers to capture the locality and discriminating information of data manifold by utilizing the local geometry information of category-specific dictionaries rather than input data. Furthermore, graph-based pairwise local constraints are designed and incorporated into the original dictionary learning model to effectively encode the locality consistency with intra-class samples and the locality inconsistency with inter-class samples. Specifically, we learn the discriminative localities for representations by jointly optimizing both the intra-class locality and inter-class locality, which can significantly improve the discriminability and robustness of dictionary. Extensive experiments on the challenging datasets verify that the proposed PLCDDL algorithm can achieve a better classification accuracy and powerful robustness compared with the state-of-the-art dictionary learning methods.

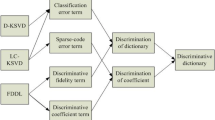
The proposed PLCDDL algorithm. 1) A pair of graph Laplacian matrices are first learned based on the class-specific dictionaries. 2) Graph-based pairwise local constraints are designed to transfer the locality for coding coefficients. 3) Class-specific dictionaries can be further updated.







Similar content being viewed by others
References
Sevakula RK, Singh V, Verma NK et al (2018) Transfer learning for molecular cancer classification using deep neural networks. IEEE ACM T Comput Bi 16(6):2089–2100
Wan T, Zhang W, Zhu M, Chen J, Achim A, Qin Z (2017) Automated mitosis detection in histopathology based on non-gaussian modeling of complex wavelet coefficients. Neurocomputing. 237:291–303
Lu C, Ma Z, Mandal M (2015) Automated segmentation of the epidermis area in skin whole slide histopathological images. IET Image Process 9(9):735–742
Kumar N, Verma R, Sharma S et al (2017) A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imag 36(7):1550–1560
Li X, Zhang X, Ding M (2019) A sum-modified-Laplacian and sparse representation based multimodal medical image fusion in Laplacian pyramid domain. Medical & biological engineering & computing 57(10):2265–2275
Huang P, Lee C (2009) Automatic classification for pathological prostate images based on fractal analysis. IEEE Trans Med Imag 28(7):1037–1050
Peyret R, Bouridane A, Khelifi F, Tahir MA, al-Maadeed S (2018) Automatic classification of colorectal and prostatic histologic tumor images using multiscale multispectral local binary pattern texture features and stacked generalization. Neurocomputing. 275:83–93
Song Y, Cai W, Zhou Y et al (2013) Feature-based image patch approximation for lung tissue classification. IEEE Trans Med Imag 32(4):797–808
Rathore S, Hussain M, Iftikhar M et al (2014) Ensemble classification of colon biopsy images based on information rich hybrid features. Comput Biol Med 47:76–92
Christodoulou C, Pattichis C, Pantziaris M et al (2003) Texture-based classification of atherosclerotic carotid plaques. IEEE Trans Med Imag. 22(7):902–912
Doyle S, Feldman M, Tomaszewski J, Madabhushi A (2012) A boosted Bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans Biomed Eng 59(5):1205–1218
Gorelick L, Veksler O, Gaed M, Gomez JA, Moussa M, Bauman G, Fenster A, Ward AD (2013) Prostate histopathology: learning tissue component histograms for cancer detection and classification. IEEE Trans Med Imag. 32(10):1804–1818
Paul A, Mukherjee D, Das P et al (2018) Improved random forest for classification. IEEE Trans Image Process 27(8):4012–4024
Wright J, Yang A, Ganesh A et al (2009) Robust face recognition via sparse representation. IEEE Trans Pattern Anal Mach Intell 31(2):210–227
Srinivas U, Mousavi H, Jeon C et al (2013) SHIRC: A simultaneous sparsity model for histopathological image representation and classification. In: IEEE 10th International Symposium on Biomedical Imagin, pp 1118–1121
Srinivas U, Mousavi H, Monga V et al (2014) Simultaneous sparsity model for histopathological image representation and classification. IEEE Trans Med Imag 33(5):1163–1179
Chang H, Nayak N, Spellman P et al (2013) Characterization of tissue histopathology via predictive sparse decomposition and spatial pyramid matching. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp 91–98
Zhou Y, Chang H, Barner K et al (2014) Classification of histology sections via multispectral convolutional sparse coding. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 3081–3088
Chang H, Han J, Zhong C, Snijders AM, Mao JH (2017) Unsupervised transfer learning via multi-scale convolutional sparse coding for biomedical applications. IEEE Trans Pattern Anal Mach Intell 40(5):1182–1194
Tang Q, Liu Y, Liu H et al (2017) Medical image classification via multiscale representation learning. Artif Intell Med 79:71–78
Zhang X., Dou H., Ju T., Xu J., Zhang S.: ‘Fusing heterogeneous features from stacked sparse autoencoder for histopathological image analysis’, IEEE J Biomed Health Inform, 2016, 20(5), pp:1377–1383
Zhang Q, Li B (2010) Discriminative K-SVD for dictionary learning in face recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 2691–2698
Jiang Z, Lin Z, Davis L et al (2013) Label consistent k-svd: learning a discriminative dictionary for recognition. IEEE Trans Pattern Anal Mach Intell 35(11):2651–2664
Yang M, Zhang L, Feng X et al (2011) Fisher discrimination dictionary learning for sparse representation. In: International Conference on Computer Vision. IEEE, Piscataway, pp 543–550
Gao S, Tsang I, Ma Y et al (2014) Learning category-specific dictionary and shared dictionary for fine-grained image categorization. IEEE Trans Image Process 23(2):623–634
Zhang R, Shen J, Wei F, Li X, Sangaiah AK (2017) Medical image classification based on multi-scale non-negative sparse coding. Artif Intell Med 83:44–51
Vu T, Mousavi H, Monga V et al (2016) Histopathological image classification using discriminative feature-oriented dictionary learning. IEEE Trans Med Imag. 35(3):738–751
Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S (2017) Breast cancer multi-classification from histopathological images with structured deep learning model. Sci Rep 7(1):4172–4182
Xu Y, Jia Z, Wang L et al (2017) Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics 18(1):281–298
Shi X, Sapkota M, Xing F, Liu F, Cui L, Yang L (2018) Pairwise based deep ranking hashing for histopathology image classification and retrieval. Pattern Recog 81:14–22
Belkin M, Niyogi P (2003) Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput 15(6):1373–1396
Wang J, Yang J, Yu K et al (2010) Locality-constrained linear coding for image classification. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 3360–3367
Zheng M, Bu J, Chen C et al (2011) Graph regularized sparse coding for image representation. IEEE Trans Image Process 20(5):1327–1336
Zhang Z, Zhao M, Chow T et al (2015) Graph based constrained semi-supervised learning framework via label propagation over adaptive neighborhood. IEEE Trans Knowl Data 27(9):362–2376
Min H, Liang M, Luo R, Zhu J (2016) Laplacian regularized locality-constrained coding for image classification. Neurocomputing. 171:1486–1495
Li Z, Lai Z, Xu Y, Yang J, Zhang D (2017) A locality-constrained and label embedding dictionary learning algorithm for image classification. IEEE Trans Neural Netw Learn Syst 28(2):278–293
Yuan Y, Li B, Meng M et al (2017) WCE abnormality detection based on saliency and adaptive locality-constrained linear coding. IEEE Trans Autom Sci Eng 14(1):149–159
Zhu X, Suk H, Huang H et al (2017) Low-rank graph-regularized structured sparse regression for identifying genetic biomarkers. IEEE Trans Big Data 3(4):405–414
Sadeghi M, Babaie M, Jutten C et al (2014) ‘Learning overcomplete dictionaries based on atom-by-atom updating’, IEEE Trans. Signal Process 62(4):883–891
Tropp J, Gilbert A et al (2007) Signal recovery from random measurements via orthogonal matching pursuit. IEEE Trans Inf Theory 53(12):4655–4666
Monga V (2013) Animal diagnostics lab dataset [Online]. Available http://signal.ee.psu.edu/medical_imaging.html
Spanhol F, Oliveira L, Petitjean C et al (2016) A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng 63(7):1455–1462
Acknowledgments
We would especially like to thank Associate Professor Chaoyang Ai for his contribution to the English revision of this manuscript.
Funding
This article is supported by Science and Technology Plan Project of Hunan Province in China (2016TP1020), Open fund project of Hunan Provincial Key Laboratory of Intelligent Information Processing and Application for Hengyang normal university, Natural Science Foundation of Hunan Province in China (2020JJ4588, 2020JJ4090).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tang, H., Mao, L., Zeng, S. et al. Discriminative dictionary learning algorithm with pairwise local constraints for histopathological image classification. Med Biol Eng Comput 59, 153–164 (2021). https://doi.org/10.1007/s11517-020-02281-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-020-02281-y