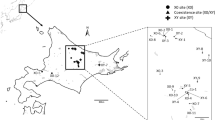
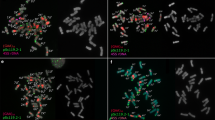
Abstract
Evolution of polyploid Aegilops species constituting complex Crassa was studied using C-banding and fluorescence in situ hybridization (FISH) with ten DNA probes. Genetic classification of nine of 14 chromosomes was suggested based on hybridization patterns of the D-genome specific repeats and distribution of rDNA loci; homoeologous groups and genome affinity of five chromosomes remain unknown. Correspondence between C-banding and FISH patterns of the repetitive DNAs was established with the aid of GAAn probe. We confirmed that the D1 genome of Ae. crassa (2n = 4x = 28, D1D1XcrXcr) was contributed by Ae. tauschii, although the retention of minor NORs on chromosomes 1D1 and 6D1 indicated that Ae. crassa probably emerged prior to the loss of the respective loci in the diploid progenitor. The Xcr genome might originate from the ancestral S*-genome species of Emarginata group. Genomes of Ae. crassa (4x) were significantly modified during speciation. The translocation Acr-6Xcr (T1) was identified in one tetraploid and all hexaploid Ae. crassa accessions and in Ae. vavilovii. Aegilops crassa (6x) also possessed the species-specific translocation 4D1-Fcr. Aegilops vavilovii originated from hybridization of Ae. crassa (4x) with Ae. searsii and was characterized by high translocation polymorphism. Two karyotypic groups: juv-I and juv-II were identified in Ae. juvenalis. All genomes of juv-I were significantly modified, whereas juv-II was karyotypically similar to Ae. crassa (4x) and Ae. umbellulata. Probably, juv-II originated independently of juv-I, from more recent hybridization of the same parental species.









Similar content being viewed by others
References
Abdolmalaki Z, Mirzaghaderi G, Mason AS, Badaeva ED (2019) Molecular cytogenetic analysis reveals evolutionary relationships between polyploid Aegilops species. Pl Syst Evol 305:459–475. https://doi.org/10.1007/s00606-019-01585-3
Appels R, Gerlach WL, Dennis ES, Swift H, Peacock WJ (1980) Molecular and chromosomal organization of DNA sequences coding for the ribosomal RNAs in cereals. Chromosoma 78:293–311. https://doi.org/10.1007/BF00327389
Badaeva ED, Badaev NS, Gill BS, Filatenko AA (1994) Intraspecific karyotype divergence in Triticum araraticum (Poaceae). Pl Syst Evol 192:117–145. https://doi.org/10.1007/BF00985912
Badaeva ED, Friebe B, Gill BS (1996a) Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome 39:293–306. https://doi.org/10.1139/g96-040
Badaeva ED, Friebe B, Gill BS (1996b) Genome differentiation in Aegilops. 2. Physical mapping of 5S and 18S-26S ribosomal RNA gene families in diploid species. Genome 39:1150–1158. https://doi.org/10.1139/g96-145
Badaeva ED, Friebe B, Zoshchuk SA, Zelenin AV, Gill BS (1998) Molecular cytogenetic analysis of tetraploid and hexaploid Aegilops crassa. Chrom Res 6:629–637. https://doi.org/10.1023/A:1009257527391
Badaeva ED, Amosova AV, Muravenko OV, Samatadze TE, Chikida NN, Zelenin AV, Friebe B, Gill BS (2002) Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Pl Syst Evol 231:163–190. https://doi.org/10.1007/s006060200018
Badaeva ED, Amosova AV, Samatadze TE, Zoshchuk SA, Shostak NG, Chikida NN, Zelenin AV, Raupp WJ, Friebe B, Gill BS (2004) Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Pl Syst Evol 246:45–76. https://doi.org/10.1007/s00606-003-0072-4
Badaeva E, Dedkova O, Koenig J, Bernard S, Bernard M (2008) Analysis of introgression of Aegilops ventricosa Tausch. genetic material in a common wheat background using C-banding. Theor Appl Genet 117:803–811. https://doi.org/10.1007/s00122-008-0821-4
Badaeva E, Zoshchuk S, Paux E, Gay G, Zoshchuk N, Roger D, Zelenin A, Bernard M, Feuillet C (2010) Fat element—a new marker for chromosome and genome analysis in the Triticeae. Chrom Res 18:697–709. https://doi.org/10.1007/s10577-010-9151-x
Badaeva E, Dedkova O, Zoshchuk S, Amosova A, Reader S, Bernard M, Zelenin A (2011) Comparative analysis of the N-genome in diploid and polyploid Aegilops species. Chrom Res 19:541–548. https://doi.org/10.1007/s10577-011-9211-x
Badaeva ED, Amosova AV, Goncharov NP, Macas J, Ruban AS, Grechishnikova IV, Zoshchuk SA, Houben A (2015) A set of cytogenetic markers allows the precise identification of all A-genome chromosomes in diploid and polyploid wheat. Cytogenet Genome Res 146:71–79. https://doi.org/10.1159/000433458
Badaeva ED, Ruban AS, Aliyeva-Schnorr L, Aliyeva-Schnorr L, Municio C, Hesse S, Houben A (2017) In situ hybridization to plant chromosomes. In: Liehr T (ed) Fluorescence in situ hybridization (FISH) application guide. Springer, Berlin. https://doi.org/10.1007/978-3-662-52959-1
Badaeva ED, Fisenko AV, Surzhikov SA, Yankovskaya AA, Chikida NN, Zoshchuk SA, Belousova MK, Dragovich AY (2019) Genetic heterogeneity of a diploid grass Aegilops tauschii revealed by chromosome banding methods and electrophoretic analysis of the seed storage proteins (gliadins). Russ J Genet 55:1315–1329. https://doi.org/10.1134/S1022795419110024
Bardsley D, Cuadrado A, Jack P, Harrison G, Castilho A, Heslop-Harrison JS (1999) Chromosome markers in the tetraploid wheat Aegilops ventricosa analyzed by in situ hybridization. Theor Appl Genet 99:300–304. https://doi.org/10.1007/s001220051236
Bedbrook RJ, Jones J, O’Dell M, Thompson RJ, Flavell RB (1980) A molecular description of telomeric heterochromatin in Secale species. Cell 19:545–560. https://doi.org/10.1016/0092-8674(80)90529-2
Bordbar F, Rahiminejad MR (2010) A taxonomic revision of Aegilops crassa Boiss. (Poaceae) in Iran. Iranian J Bot 16:30–35
Cabi E, Doğan M, Özler H, Akaydin G, Karagöz A (2010) Taxonomy, morphology and palynology of Aegilops vavilovii (Zhuk.) Chennav.(Poaceae: Triticeae). African J Agric Res 520:2841–2849. https://doi.org/10.5897/AJAR.9000672
Chapman V, Miller TE (1978) The relationship of the D genomes of hexaploid Ae. crassa, Ae. vavilovii and hexaploid wheat. Wheat Inform Serv 47:17–20
Chennaveeraiah MS (1960) Karyomorphologic and cytotaxonomic studies in Aegilops. Acta Horti Gotoburgensis 23:85–186
Cuadrado A, Cardoso M, Jouve N (2008) Increasing the physical markers of wheat chromosomes using SSRs as FISH probes. Genome 51:809–815. https://doi.org/10.1139/G08-065
Danilova TV, Friebe B, Gill BS (2014) Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet 127:715–730. https://doi.org/10.1007/s00122-013-2253-z
Danilova TV, Akhunova AR, Akhunov ED, Friebe B, Gill BS (2017) Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae). Pl J 92:317–330. https://doi.org/10.1111/tpj.13657
Dubcovsky J, Dvořák J (1995a) Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140:1367–1377. http://www.genetics.org/content/140/4/1367.abstract
Dubkovsky J, Dvořák J (1995) Genome identification of the Triticum crassum complex (Poaceae) with the restriction patterns of repeated nucleotide sequences. Amer J Bot 182:131–140. https://doi.org/10.2307/2445795
Dvořák J, Zhang H-B, Kota RS, Lassner M (1989) Organization and evolution of the 5S ribosomal RNA gene family in wheat and related species. Genome 32:1003–1016. https://doi.org/10.1139/g89-545
Dvořák J (1998) Genome analysis in the Triticum-Aegilops alliance. In: Slinkard AE (ed) Proceedings of the 9th international wheat genetics symposium, 2–7 August 1998. Printcrafters Inc., Saskatoon, pp 8–11
Edet OU, Gorafi YSA, Nasuda S, Tsujimoto H (2018) DArTseq-based analysis of genomic relationships among species of tribe Triticeae. Sci Rep 8:16397. https://doi.org/10.1038/s41598-018-34811-y
Eig A (1929) Monographisch-kritische ubersicht der gattung Aegilops. Beihefte, Berlin
Friebe B, Gill BS (1996) Chromosome banding and genome analysis in diploid and cultivated polyploid wheats. In: Jauhar PP (ed) Methods in genome analysis in plants: their merits and pitfalls. CRC Press, Boca Ration, pp 39–60
Friebe B, Tuleen N, Jiang J, Gill BS (1993) Standard karyotype of Triticum longissimum and its cytogenetic relationship with T. aestivum. Genome 36:731–742. https://doi.org/10.1139/g93-098
Friebe B, Jiang J, Tuleen N, Gill BS (1995a) Standard karyotype of Triticum umbellulatum and the characterization of derived chromosome addition and translocation lines in common wheat. Theor Appl Genet 90:150–156. https://doi.org/10.1007/BF00221010
Friebe B, Tuleen NA, Gill BS (1995b) Standard karyotype of Triticum searsii and its relationship with other S-genome species and common wheat. Theor Appl Genet 91:248–254. https://doi.org/10.1007/BF00220885
Friebe B, Badaeva ED, Hammer K, Gill BS (1996) Standard karyotypes of Aegilops uniaristata, Ae. mutica, Ae. comosa subspecies comosa and heldreichii (Poaceae). Pl Syst Evol 202:199–210. https://doi.org/10.1007/BF00983382
Friebe B, Qi LL, Nasuda S, Zhang P, Tuleen NA, Gill BS (2000) Development of a complete set of Triticum aestivum-Aegilops speltoides chromosome addition lines. Theor Appl Genet 101:51–58. https://doi.org/10.1007/s001220051448
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucl Acids Res 7:1869–1885. https://doi.org/10.1093/nar/7.7.1869
Gerlach WL, Dyer TA (1980) Sequence organization of the repeated units in the nucleus of wheat which contains 5S-rRNA genes. Nucl Acids Res 8:4851–4865. https://doi.org/10.1007/BF00327389
Hamid HM, Habibollah S, Rasool AZ, Nasrin M, Esmaeil FS (2010) A comparative study of chromosome morphology among some accessions of Aegilops crassa. African J Biotechnol 9:996–1000. https://doi.org/10.5897/AJB09.243
Hart GE, Tuleen NA (1983) Introduction and characterization of alien genetic material. In: Tanksley SD, Orton TJ (eds) Isozymes in plant genetics and breeding part A. Elsevier Science Publishers B.V, Amsterdam, Oxford, New York, pp 339–362
Jiang J, Gill BS (1994) New 18S-26S ribosomal RNA gene loci: chromosomal landmarks for the evolution of polyploid wheats. Chromosoma 103:179–185. https://doi.org/10.1007/BF00368010
Jovkova ME, Kondeva E, Kostova R (1977) Biochemical investigations on Aegilops crassa x Triticum aestivum hybrids. Genet Selekts 10:91–98
Kihara H (1949) Genomanalyse bei Triticum und Aegilops. IX. Systematischer Aufbau der Gattung Aegilops auf genomanalytischer Grundlage. Cytologia 14:135–144. https://doi.org/10.1508/cytologia.14.135
Kihara H (1954) Considerations on the evolution and distribution of Aegilops species based on the analyzer-method. Cytologia 19:336–357. https://doi.org/10.1508/cytologia.19.336
Kihara H (1957) Completion of genome-analysis of three 6x species of Aegilops. Wheat Inform Serv 6:11
Kihara H (1963) Interspecific relationship in Triticum and Aegilops. Seiken Ziho 15:1–12
Kihara H, Tanaka M (1970) Attendum to the classification of the genus Aegilops by means of genome analysis. Wheat Inform Serv 30:1–2
Kihara H, Yamashita K, Tanaka M, Sakamoto S (1957) Geographical distribution of 4x and 6x forms of Aegilops crassa. Wheat Inform Serv 5:11
Kihara H, Yamashita K, Tanaka M (1959) Genomes of 6x species of Aegilops. Wheat Inform Serv 8:3–5
Kilian B, Mammen K, Millet E, Sharma R, Graner A, Salamini F, Hammer K, Ozkan H (2011) Aegilops. In: Kole C (ed) Wild crop relatives: genomics and breeding resources Cereals. Springer, Berlin, pp 1–76. https://doi.org/10.1007/978-3-642-14228-4
Kimber G, Abubakar M (1979) Wheat hybrid information system. Cereal Res Commun 7:257–280. https://www.jstor.org/stable/23779262
Kimber G, Feldman M (1987) Wild wheat, an introduction. College of Agriculture University of Missouri, Columbia
Kimber G, Sears ER (1984) Assignment of genome symbols in the Triticeae. In: Sakamoto S (ed) Proceedings of the 6th international wheat genetics symposium, November 28–December 3 1983. Plant Germ-Plasm Institute, Kyoto University, Kyoto, pp 1195–1196
Kimber G, Tsunewaki K (1988) Genome symbols and plasma types in the wheat group. In: Miller TE, Koebner RMD (eds) Proceedings of the 7th international wheat genetics symposium, 13–19 July 1988. Bath Press, Avon, Cambridge, pp 1209–1210
Kimber G, Yen Y (1989) Hybrids between wheat relatives and autotetraploid Triticum umbellulatum. Genome 32:1–5. https://doi.org/10.1139/g89-401
Kimber G, Zhao YH (1983) The D genome of the Triticeae. Canad J Genet Cytol 25:581–589. https://doi.org/10.1139/g83-088
Kishii M (2019) An update of recent use of Aegilops species in wheat breeding. Frontiers Pl Sci 10:585. https://doi.org/10.3389/fpls.2019.00585
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137. https://doi.org/10.1139/gen-2013-0003
Kumar A, Kapoor P, Chunduri V, Sharma S, Garg M (2019) Potential of Aegilops sp. for improvement of grain processing and nutritional quality in wheat (Triticum aestivum). Frontiers Pl Sci 10:308. https://doi.org/10.3389/fpls.2019.00308
Li C, Sun X, Conover JL, Zhang Z, Wang J, Wang X, Deng X, Wang H, Liu B, Wendel JF, Gong L (2018) Cytonuclear coevolution following homoploid hybrid speciation in Aegilops tauschii. Molec Biol Evol 36:341–349. https://doi.org/10.1093/molbev/msy215
Lilienfeld FA (1951) H. Kihara: Genome-analysis in Triticum and Aegilops X. Concluding review. Cytologia 16:101–123
Liu C, Gong W, Han R, Guo J, Li G, Li H, Song J, Liu A, Cao X, Zhai S, Cheng D, Li G, Zhao Z, Yang Z, Liu J, Reader SM (2019) Characterization, identification and evaluation of a set of wheat-Aegilops comosa chromosome lines. Sci Rep 9:4773. https://doi.org/10.1038/s41598-019-41219-9
Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, Jakobsen KS, Wulff BBH, Steuernagel B, Mayer KFX, Olsen O-A (2014) Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092. https://doi.org/10.1126/science.1250092
McGinnis RC, Melnyk JH (1956) Cytological evidence for the S genome in Aegilops juvenalis. Wheat Inform Serv 4:8
Migushova EP, Khakimova AG (1982) To the questions of systematics of Aegilops crassa Boiss. Bull VIR 119:75–76 (in Russian)
Molnár I, Vrána J, Burešová V, Cápal P, Farkas A, Darkó É, Cseh A, Kubaláková M, Molnár-Láng M, Doležel J (2016) Dissecting the U, M, S and C genomes of wild relatives of bread wheat (Aegilops spp.) into chromosomes and exploring their synteny with wheat. Pl J 88:452–467. https://doi.org/10.1111/tpj.13266
Molnár-Láng M, Molnár I, Szakács É, Linc G, Bedö Z (2014) Production and molecular cytogenetic identification of wheat-alien hybrids and introgression lines. In: Tuberosa R, Graner A, Frison E (eds) Genomics of plant genetic resources, vol. 1. Managing, sequencing and mining genetic resources. Springer, New York, Heidelberg, Dordrecht, London, pp 255–284
Molnár-Láng M, Ceoloni C, Doležel J (2015) Alien introgression in wheat cytogenetics, molecular biology, and genomics. Springer, Cham
Monneveux P, Zaharieva M, Rekika D (2000) The utilization of Triticum and Aegilops species for the improvement of durum wheat. In: Royo C, Nachit M, Di Fonzo N, Araus JL (eds) Durum wheat improvement in the Mediterranean region: new challenges. CIHEAM, Zaragoza, pp 71–81
Morris R, Sears ER (1967) The cytogenetics of wheat and its relatives. In: Quisenberry RS, Reitz LP (eds) Wheat and Wheat Improvement. American Society of Agronomy, Madison, pp 19–87
Mukai Y, Endo TR, Gill BS (1991) Physical mapping of the 18S.26S rRNA multigene family in common wheat: identification of a new locus. Chromosoma 100:71–78. https://doi.org/10.1007/BF00418239
Nakai Y (1982) D genome donors for Aegilops crassa (DDMM, DDDDMM) and Ae. vavilovii (DDMMSS) deduced from esterase analysis by isoelectric focusing. Jap J Genet 57:349–360
Ogihara Y, Tsunewaki K (1988) Diversity and evolution of chloroplast DNA in Triticum and Aegilops as revealed by restriction fragment analysis. Theor Appl Genet 76:321–332. https://doi.org/10.1007/BF00265331
Olivera PD, Rouse MN, Jin Y (2018) Identification of new sources of resistance to wheat stem rust in Aegilops spp. in the tertiary genepool of wheat. Frontiers Pl Sci 9:1719. https://doi.org/10.3389/fpls.2018.01719
Parisod C, Badaeva ED (2020) Chromosome restructuring among hybridizing wild wheats. New Phytol 226:1263–1273. https://doi.org/10.1111/nph.16415
Pedersen C, Langridge P (1997) Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome 40:589–593. https://doi.org/10.1139/gen-40-5-589
Przewieslik-Allen AM, Burridge AJ, Wilkinson PA, Winfield MO, Shaw DS, McAusland L, King J, King IP, Edwards KJ, Barker GLA (2019) Developing a high-throughput SNP-based marker system to facilitate the introgression of traits from Aegilops species into bread wheat (Triticum aestivum). Frontiers Pl Sci 9:1993. https://doi.org/10.3389/fpls.2018.01993
Rather S, Sharma D, Pandey I, Joshi N (2017) Alien gene introgression in wheat. In: Kumar A, Kumar A, Prasad B (eds) Wheat a premier food crop. Kalyani, India, pp 101–120
Rayburn AL, Gill BS (1986) Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Pl Molec Biol Rep 4:102–109. https://doi.org/10.1007/BF02732107
Rayburn AL, Gill BS (1987) Molecular analysis of the D-genome of the Triticeae. Theor Appl Genet 73:385–388. https://doi.org/10.1007/BF00262505
Ruban AS, Badaeva ED (2018) Evolution of the S-genomes in Triticum-Aegilops alliance: evidences from chromosome analysis. Frontiers Pl Sci 9:1756. https://doi.org/10.3389/fpls.2018.01756
Schneider A, Molnár I, Molnár-Láng M (2008) Utilization of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163:1–19. https://doi.org/10.1007/s10681-007-9624-y
Talbert LE, Magyar GM, Lavin M, Moylan SL, Blake TK (1991) Molecular evidence for the origin of S-derived genomes of polyploid Triticum species. Amer J Bot 78:340–349. https://doi.org/10.2307/2444956
Tang Z, Yang Z, Fu S (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318. https://doi.org/10.1007/s13353-014-0215-z
Tang S, Qiu L, Xiao Z, Fu S, Tang Z (2016) New oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 7:118. https://doi.org/10.3390/genes7120118
Tsunewaki K (1996) Plasmon analysis as the counterpart of genome analysis. In: Jauhar PP (ed) Methods of genome analysis in plant: their merits and pitfalls. CRC Press, Boca Ration, pp 271–299
Van Slageren MW (1994) Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. et Spach) Eig (Poaceae). Wageningen Agricultural University and ICARDA, Wageningen and Aleppo
Wang K, Guo W, Zhang T (2007) Detection and mapping of homologous and homoeologous segments in homoeologous groups of allotetraploid cotton by BAC-FISH. BMC Genom 8:1–8. https://doi.org/10.1186/1471-2164-8-178
Witcombe JR (1983) A guide to the species of Aegilops L. Their taxonomy, morphology and distribution. IBPGR Secretariat, Rome
Xu S, Dong Y (1992) Fertility and meiotic mechanisms of hybrids between chromosome autoduplication tetraploid wheats and Aegilops species. Genome 35:379–384. https://doi.org/10.1139/g92-057
Yamamoto M (1992a) Detection of ribosomal RNA genes in Aegilops by in situ hybridization. Bull Osaka Private Coll Assoc 29:77–82
Yamamoto M (1992b) Distribution of ribosomal RNA genes in Aegilops and Triticum chromosomes. Bull Kansai Women’s Coll 2:25–37
Yen Y, Kimber G (1992) The S genome in Triticum syriacum. Genome 35:709–713. https://doi.org/10.1139/g92-109
Zhang HB, Dvořák J (1992) The genome origin and evolution of hexaploid Triticum crassum and Triticum syriacum determined from variation in repeated nucleotide sequences. Genome 35:806–814. https://doi.org/10.1139/g92-123
Zhang P, Dundas IS, McIntosh RA, Xu SS, Park RF, Gill BS, Friebe B (2015) Wheat– Aegilops introgressions. In: Molnár-Láng M, Ceoloni C, Doležel J (eds) Alien introgression in wheat cytogenetics, molecular biology, and genomics. Springer, Cham, pp 221–244
Zhao YH, Kimber G (1984) New hybrids with D genome wheat relatives. Genetics 106:509–515. http://www.genetics.org/cgi/content/abstract/106/3/509
Zhao L, Ning S, Yi Y, Zhang L, Yuan Z, Wang J, Zheng Y, Hao M, Liu D (2018) Fluorescence in situ hybridization karyotyping reveals the presence of two distinct genomes in the taxon Aegilops tauschii. BMC Genom 19:3. https://doi.org/10.1186/s12864-017-4384-0
Zhukovsky PM (1928) A critical-systematical survey of the species of the genus Aegilops L. Bull Appl Bot Genet Pl Breed 18:417–609 (in Russian)
Zohary D, Feldman M (1962) Hybridization between amphiploids and the evolution of polyploids in the wheat (Aegilops-Triticum) group. Evolution 16:44–61. https://doi.org/10.2307/2406265
Acknowledgements
The authors thank Drs. B.S. Gill (WGGRS, KSU, Kansas, USA) H. Bockelman (USDA-ARS, Aberdeen, Idaho, USA); Andreas Börner (IPK, Gatersleben, Germany), J. Valkoun (ICARDA, Aleppo, Syria) for supplying the material.
Funding
The study was supported in part by grant from the Russian State Foundation for Basic Research (projects 17-04-00087a, 20-04-00284a) and by State Budgetary Projects No 0112-2019-0002 (from VIGG) and No 0662-2019-0006 (from VIR).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. NNC and MKhB provided material for analysis, slide preparation and FISH experiments were performed by EDB, ASR and SAZ, oligo-probes were synthesized by SAS, and data analysis was performed by EDB. EDB wrote the first draught of the manuscript and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest and that accepted principles of ethical and professional conduct have been followed. The authors agree to participate in the study and submit their results for publication
Ethics approval
Not applicable.
Additional information
Handling Editor: Martin A. Lysak.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1. The list of plant materials, including genome constitution, methods of karyotype analysis, translocation types, origin and collection sites of accessions.
Online Resource 2. Characteristics of oligo-probes.
Online Resource 3. Translocation variants of polyploid Aegilops species of the complex Crassa identified using FISH with nine DNA probes.
Online Resource 4. Comparison of the C-banding patterns of Ae. juvenalis accessions juv-I (a-e) and juv-II (f-g).
Rights and permissions
About this article
Cite this article
Badaeva, E.D., Chikida, N.N., Belousova, M.K. et al. A new insight on the evolution of polyploid Aegilops species from the complex Crassa: molecular-cytogenetic analysis. Plant Syst Evol 307, 3 (2021). https://doi.org/10.1007/s00606-020-01731-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-020-01731-2