Abstract
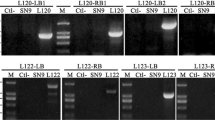
Elucidation of the genomic organizations of transgene insertion sites is essential for the genetic studies of transgenic plants. Herein, we establish an analysis pipeline that identifies the transgene insertion sites as well as the presence of vector backbones, through de novo genome assembly with high-throughput sequencing data in two transgenic soybean lines, AtYUCCA6-#5 and 35S-UGT72E3/2-#7. Sequencing data of approximately 28× and 29× genome coverages for each line generated by high-throughput sequencing were de novo assembled. The databases generated from the de novo assembled sequences were used to search contigs that contained putative insertion sites and their flanking sequences (integration sites) of transgene fragments using transgenic vector sequences as queries. The predicted integration site sequences, which are located at three annotated genes that might regulate plant development or confer disease resistance, were then confirmed by local alignment against the soybean reference genome and PCR amplification. As results, we revealed the precise transgene-flanking sequences and sequence rearrangements at insertion sites in both the transgenic lines, as well as the aberrant insertion of a transgene fragment. Consequently, relative to experimental or enrichment technologies, our approach is straightforward and time-effective, providing an alternative method for the identification of insertion sites in transgenic plants.



Similar content being viewed by others
References
Al-Babili S, Beyer P (2005) Golden Rice - five years on the road - five years to go? Trends Plant Sci 10:565–573. https://doi.org/10.1016/j.tplants.2005.10.006
Alonso JM, Stepanova AN, Leisse TJ et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 301:653–657. https://doi.org/10.1126/science.1086391
An S, Park S, Jeong DH, Lee DY, Kang HG, Yu JH, Hur J, Kim SR, Kim YH, Lee M, Han S, Kim SJ, Yang J, Kim E, Wi SJ, Chung HS, Hong JP, Choe V, Lee HK, Choi JH, Nam J, Kim SR, Park PB, Park KY, Kim WT, Choe S, Lee CB, An G (2003) Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 133:2040–2047. https://doi.org/10.1104/pp.103.030478
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Report 9:208–218
Azpiros-Leeban R, Feldmann KA (1997) T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet 13:162–156. https://doi.org/10.1016/S0168-9525(97)01094-9
Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IMJ, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DMD, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara E. Catenazzi M, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, vandeVondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59. https://doi.org/10.1038/nature07517
Cade R, Burgin K, Schilling K et al (2018) Regulatory science evaluation of whole genome sequencing and an insertion site characterization method for molecular characterization of GM maize. J Regul Sci 6:1–14
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:1–9. https://doi.org/10.1186/1471-2105-10-421
De Buck S, De Wilde C, Van Montagu M, Depicker A (2000) T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol Breed 6:459–468. https://doi.org/10.1023/A:1026575524345
Guo B, Guo Y, Hong H, Qiu LJ (2016) Identification of genomie insertion and flanking sequence of G2-EPSPS and GAT transgenes in soybean using whole genome sequencing method. Front Plant Sci 7:1–9. https://doi.org/10.3389/fpls.2016.01009
Guttikonda SK, Marri P, Mammadov J, Ye L, Soe K, Richey K, Cruse J, Zhuang M, Gao Z, Evans C, Rounsley S, Kumpatla SP (2016) Molecular characterization of transgenic events using next generation sequencing approach. PLoS One 11:e0149515. https://doi.org/10.1371/journal.pone.0149515
Jeong SC, Hayes AJ, Biyashev RM, Maroof MAS (2001) Diversity and evolution of a non-TIR-NBS sequence family that clusters to a chromosomal “hotspot” for disease resistance genes in soybean. Theor Appl Genet 103:406–414. https://doi.org/10.1007/s001220100567
Jeong S-C, Pack IS, Cho E-Y, Youk ES, Park S, Yoon WK, Kim CG, Choi YD, Kim JK, Kim HM (2007) Molecular analysis and quantitative detection of a transgenic rice line expressing a bifunctional fusion TPSP. Food Control 18:1434–1442. https://doi.org/10.1016/j.foodcont.2006.10.007
Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, Kohara Y, Fujiyama A, Hayashi T, Itoh T (2014) Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 24:1384–1395. https://doi.org/10.1101/gr.170720.113
Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, Seo E, Choi J, Cheong K, Kim KT, Jung K, Lee GW, Oh SK, Bae C, Kim SB, Lee HY, Kim SY, Kim MS, Kang BC, Jo YD, Yang HB, Jeong HJ, Kang WH, Kwon JK, Shin C, Lim JY, Park JH, Huh JH, Kim JS, Kim BD, Cohen O, Paran I, Suh MC, Lee SB, Kim YK, Shin Y, Noh SJ, Park J, Seo YS, Kwon SY, Kim HA, Park JM, Kim HJ, Choi SB, Bosland PW, Reeves G, Jo SH, Lee BW, Cho HT, Choi HS, Lee MS, Yu Y, Do Choi Y, Park BS, van Deynze A, Ashrafi H, Hill T, Kim WT, Pai HS, Ahn HK, Yeam I, Giovannoni JJ, Rose JKC, Sørensen I, Lee SJ, Kim RW, Choi IY, Choi BS, Lim JS, Lee YH, Choi D (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46:270–278. https://doi.org/10.1038/ng.2877
Kovalic D, Garnaat C, Guo L, Yan Y, Groat J, Silvanovich A, Ralston L, Huang M, Tian Q, Christian A, Cheikh N, Hjelle J, Padgette S, Bannon G (2012) The use of next generation sequencing and junction sequence analysis bioinformatics to achieve molecular characterization of crops improved through modern biotechnology. Plant Genome 5:149–163. https://doi.org/10.3835/plantgenome2012.10.0026
Kwon T, Kim HJ, Yun SY, Kim J, Cho HS, Nam J, Chung YS (2017) Enhancement of syringin contents in soybean seeds with seed-specific expression of a chimeric UGT72E3/E2 gene. Plant Biotechnol Rep 11:439–447. https://doi.org/10.1007/s11816-017-0464-5
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Leoni C, Volpicella M, De Leo F et al (2011) Genome walking in eukaryotes. FEBS J 278:3953–3977. https://doi.org/10.1111/j.1742-4658.2011.08307.x
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Merchant S, Wood DE, Salzberg SL (2014) Unexpected cross-species contamination in genome sequencing projects. PeerJ 2014:1–7. https://doi.org/10.7717/peerj.675
Park D, Park S, Ban YW et al (2017) A bioinformatics approach for identifying transgene insertion sites using whole genome sequencing data. BMC Biotechnol 17:67. https://doi.org/10.1186/s12896-017-0386-x
Park D, Park SH, Kim YS, Choi BS, Kim JK, Kim NS, Choi IY (2019a) NGS sequencing reveals that many of the genetic variations in transgenic rice plants match the variations found in natural rice population. Genes and Genomics 41:213–222. https://doi.org/10.1007/s13258-018-0754-5
Park JS, Kim HJ, Cho HS, Jung HW, Cha JY, Yun DJ, Oh SW, Chung YS (2019b) Overexpression of AtYUCCA6 in soybean crop results in reduced ROS production and increased drought tolerance. Plant Biotechnol Rep 13:161–168. https://doi.org/10.1007/s11816-019-00527-2
Pauwels K, De Keersmaecker SCJ, De Schrijver A et al (2015) Next-generation sequencing as a tool for the molecular characterisation and risk assessment of genetically modified plants: added value or not? Trends Food Sci Technol 45:319–326. https://doi.org/10.1016/j.tifs.2015.07.009
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A 81:8014–8018
Siddique K, Wei J, Li R, Zhang D, Shi J (2019) Identification of T-DNA insertion site and flanking sequence of a genetically modified maize event IE09S034 using next-generation sequencing technology. Mol Biotechnol 61:694–702. https://doi.org/10.1007/s12033-019-00196-0
Smith N, Kilpatrick JB, Whitelam GC (2001) Superfluous transgene integration in plants. CRC Crit Rev Plant Sci 20:215–249. https://doi.org/10.1080/20013591099218
Yang L, Wang C, Holst-Jensen A, Morisset D, Lin Y, Zhang D (2013) Characterization of GM events by insert knowledge adapted re-sequencing approaches. Sci Rep 3:1–9. https://doi.org/10.1038/srep02839
Funding
This work was supported by the Korea Research Institute of Bioscience and Biotechnology Research Initiative Program and partly by the National Research Foundation grant (NRF-2018R1A2A2A05021904) funded by the Korean government.
Author information
Authors and Affiliations
Contributions
S.C.J conceived the presented idea and designed the project. M.S.K performed the bioinformatics analysis. C.G.K, T.K, J.N., and Y.S.C. provided transgenic plant materials. H.J. J.H.K., D.N.B., and I.S.P. performed experiments. S.C.J. and H.J. wrote the manuscript with input from all other authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, MS., Jo, H., Kim, J.H. et al. Elucidation of genomic organizations of transgenic soybean plants through de novo genome assembly with short paired-end reads. Mol Breeding 41, 1 (2021). https://doi.org/10.1007/s11032-020-01191-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-020-01191-z