Contrasting Trends of Population Size Change for Two Eurasian Owlet Species—Athene brama and Glaucidium radiatum From South Asia Over the Late Quaternary
- 1Sálim Ali Centre for Ornithology and Natural History, Coimbatore, India
- 2Indian Institute of Science Education and Research (IISER), Tirupati, India
- 3Indian Institute of Science Education and Research (IISER), Bhopal, India
Climatic oscillations over the Quaternary have had a lasting impact on species’ distribution, evolutionary history, and genetic composition. Many species show dramatic population size changes coinciding with the last glacial period. However, the extent and direction of change vary across biogeographic regions, species-habitat associations, and species traits. Here we use genomic data to assess population size changes over the late Quaternary using the Pairwise Sequential Markovian Coalescent (PSMC) approach in two Eurasian Owlet species—the Spotted Owlet, Athene brama, and the Jungle Owlet, Glaucidium radiatum. While Spotted Owlets are typically associated with open habitats, Jungle Owlets are found in deciduous forests and scrublands. We find that the effective population size for the Spotted Owlet increased after the Interglacial period till the Last Glacial Maxima and subsequently declined toward the Mid-Holocene. On the other hand, effective population size estimates for the Jungle Owlet increased gradually throughout this period. These observations are in line with climatic niche model-based predictions for range size change for both species from a previous study and suggest that habitat associations at the local scale are important in determining responses to past climatic and vegetational changes. The Spotted Owlet result also aligns well with the expectation of open habitat expansion during the arid Glacial Maxima, whereas for the Jungle Owlet the contrasting expectation does not hold. Therefore, assessing the impacts of glacial history on population trajectories of multiple species with different habitat associations is necessary to understand the impacts of past climate on South Asian taxa.
Introduction
Quaternary climatic fluctuations, characterized by repeated cycles of cool and arid glacial periods punctuated by warm “interglacials,” are notable for their role in shaping the distribution and evolutionary history of species globally (Hewitt, 2000, 2004). Northern latitudes experienced far greater extremes with glacial periods being marked by an expansion of ice sheets, retraction of temperate and boreal forests, and expansion of tundra vegetation (Hofreiter and Stewart, 2009; Beyer et al., 2020). Lowered sea levels exposed a greater land area and created land bridges. Though less pronounced, the tropics too experienced changes in temperature and precipitation, with glacial periods being drier and cooler, leading to contraction and fragmentation of tropical forests, expansion of grasslands, or dry woodlands (Qian and Ricklefs, 2001; Ray and Adams, 2001). Coincidental with these shifts in vegetation and available habitat, species ranges changed dynamically through the Quaternary, varying in their extent and geographical location (Dynesius and Jansson, 2000), impacting community composition and species distribution (Fuchs et al., 2019). Along with this, changes in population size and isolation-migration patterns altered the genetic composition of populations (Hewitt, 2000).
In the tropics, there is considerable empirical support for the impact of forest fragmentation and open habitat expansion during the glacial periods on the population history of many species (Schneider and Moritz, 1999; Cabanne et al., 2007; Wurster et al., 2010; Piñeiro et al., 2017; Song et al., 2020). However, data contradicting this general pattern (Batalha-Filho et al., 2012; Rocha and Kaefer, 2019) underscore the importance of understanding regional patterns across taxonomic groups. Among tropical regions, South Asia remains poorly represented in studies that include genetic data (Reddy, 2014), even fewer that examine historical climatic oscillation-induced species range dynamics. Global reconstructions of vegetation indicate that the region saw an expansion of open habitats including grasslands and dry scrubland during the last glacial period (Beyer et al., 2020). Palynological and isotope analysis suggest that monsoonal activity was also reduced during this period while being stronger during the interglacial (Prabhu et al., 2004). These changes are likely to have impacted forest and open-habitat associated species in an opposing manner, but to varying degrees depending on the strengths of association with the habitat.
Here we examine the impact of the late Quaternary climatic events on two phylogenetically close (from the same family—Strigidae) but ecologically differentiated avian species from the Indian subcontinent. The Spotted Owlet, Athene brama and the Jungle Owlet, Glaucidium radiatum, are small owls, widely distributed, and sympatric over large parts of their current distribution. Nevertheless, they have distinct dietary habits and breeding seasons (Mehta et al., 2018), and despite their range overlap, at smaller spatial scales they are segregated by their habitat preferences. Spotted Owlets avoid dense forests and are primarily associated with more open habitats such as cultivated areas, semi-desert areas, and open woodlands (Ali and Ripley, 1983; Holt et al., 2020b). Jungle Owlets are typically found in forested areas, including dense and secondary dry/moist deciduous forests, scrublands, and riparian areas (Ali and Ripley, 1983; Holt et al., 2020a). Therefore, it is likely that suitable habitats for the two species expanded and contracted in opposition to each other, with the glacial periods being favorable for Spotted Owlets and the interglacials for Jungle Owlets. Genomic data allow us to assess such predictions by looking for temporal changes in effective population size (Ne) coinciding with major climatic events.
The Pairwise Sequentially Markovian Coalescent (PSMC) model has frequently been used to infer changes in Ne in the context of past climate (Nadachowska-Brzyska et al., 2015; Vijay et al., 2018; Lucena-Perez et al., 2020). This model relies on the rate of coalescence along a single diploid genome and uses this to infer population size over time (Li and Durbin, 2012). The PSMC framework has been particularly useful in cases where acquiring large genomic datasets is still logistically prohibitive (Mays et al., 2018). Based on PSMC curves, several species show dynamic changes in Ne during the past glacial cycles, suggesting a strong impact of glacial cycles on population history (Nadachowska-Brzyska et al., 2015; Kozma et al., 2016; Mays et al., 2018; Vijay et al., 2018; Feng et al., 2019). Such analyses have often been used to complement and corroborate evidence from climatic niche models (Kozma et al., 2018; Mays et al., 2018). Moreover, long-term Ne trends help understand contemporary genetic variation (Xue et al., 2015) and extinction risk (Hung et al., 2014), thus providing an evolutionary context for current population trends.
Objectives
Here we test the hypothesis that the demographic history of species will be strongly affected by the ecological differences, specifically habitat preferences. We expect to see an increase in Ne from LIG-LGM followed by a decline in the LGM-MDH period for the Spotted Owlet. We expect the reverse trend for the Jungle Owlet, with contraction in Ne during the glacial period and expansion toward the Holocene. However, a previous study using climatic niche models suggests that the extent of Jungle Owlet habitat was reduced (relative to the Spotted Owlet) during the LIG and gradually increased toward the MDH (Koparde et al., 2019). We use the PSMC-based reconstruction of past Ne from whole-genome sequence data to investigate past population size dynamics of the Spotted Owlet and the Jungle Owlet over the late Quaternary.
Materials and Methods
Sample Collection
We obtained one sample per species from Central (Jungle Owlet) and Southern (Spotted Owlet) Western Ghats, with due permits from the Forest Department and extracted DNA using the DNeasy Blood and Tissue kit following the manufacturer’s protocol with slight modifications. Whole DNA concentration was quantified using a Qubit 4.0 fluorometer and DNA integrity was visualized using 1% Agarose Gel electrophoresis and Tapestation. Samples were sequenced on an Illumina HiSeq 10X platform (150 bp paired-end), targeting 30x genome coverage.
Read Mapping and Variant Calling
Raw sequences were inspected for overall quality, adapter content, and the number of reads to ensure that it met the required quality standards using the program TrimGalore!1. Reads were processed using the GATK short reads pipeline (McKenna et al., 2010) through which read-group information was added and Illumina adapters were marked. For both study species, no reference assembly was available. Therefore, the Burrowing Owl (Athene cunicularia) assembly was used as a reference genome (RAG1 nuclear gene p-distance—the proportion of nucleotide sites at which the compared sequences are different—with A. brama 1.07%; with G. radiatum 2.25%; Supplementary Table S1.). The unmasked assembly (athCun1) was downloaded from Ensembl (Yates et al., 2020). Reads were mapped against the reference using the Burrows-Wheeler Aligner (Li and Durbin, 2009; Li, 2013) with the “mem” option under default settings. PCR duplicates were marked using Picard Tools2. At this stage, the average depth and genome coverage for each sample were summarized using the “Depth Of Coverage” option from GATK as a quality check before proceeding further. The Burrowing Owl assembly has an accompanying sequence report which lists chromosomal information for each scaffold and identifies them as occurring on autosomes, the Z chromosome, or being unplaced. Therefore, reads mapping to the 51 autosomal scaffolds from 28 chromosomes were selected. The scaffold N50 of the reference genome is 42,147,404 bp (42 Mb) and the shortest scaffold used was 121,167 bp (0.12 Mb) long, making it suitable for use with PSMC (Patton et al., 2019). A bed file identifying autosomal scaffolds was created and used to extract reads aligning only to autosomes using the program samtools (Li, 2011) with the view option, along with -b -L flags. The aligned file was filtered by setting a base quality threshold (q 20). Variants were identified using the bcftools (Li, 2011) mpileup (with the -C50 flag) and call (with the -c flag) options and they were converted to a consensus file with depth filters (minimum 10, maximum 100), as suggested in the PSMC manual (Li and Durbin, 2012). The bcftools/samtools variant-calling pipeline was used, as suggested in the PSMC manual. Genome coverage was calculated using samtools depth for different depth filters to see how the filtering impacted the data.
Demographic History Through PSMC
The consensus file was converted to a suitable input format for the PSMC pipeline following instructions in the PSMC manual. PSMC analysis was repeated with different values of the “-t” and “-r” parameters, controlling the maximum time to the most recent common ancestor (TMRCA) and the number of free atomic time intervals, respectively. For each value of “-t” (starting with 15), “-p” was modified till each time span was supported by at least 20 events after 25 iterations. The final set of parameters was chosen to be “-N25 -t9 -r5 -p ‘26∗2 + 4 + 7 + 1’.” Bootstrapping with 100 replicates was performed with the identified set of parameters using a block size of 0.5 Mb. The output was written to a text format (using the -R option with the psmc_plot.pl script) for plotting in R. Estimates of the mutation rate and the generation time are required to convert the scaled population size (lambda) to effective population size (Ne) and the time (in generations) to units of years. As the mutation rate for the study species is unknown, it was assumed to be 4 × 10–9, based on an average estimate of mutation rates from chickens to passerines (Mueller et al., 2018). Nevertheless, additional plots with alternative values of mutation rate based on estimates from other species were also made for comparison. Generation time was assumed to be 2 years, calculated as the age of sexual maturity/reproduction (1 year; Pande et al., 2007) multiplied by 2, as suggested by Nadachowska-Brzyska et al. (2015). The scripts used have been uploaded to GitHub3.
Results
The Jungle Owlet and Spotted Owlet samples yielded 52.8 and 44.5 Giga bases, respectively (from 150 bp paired-end reads). After alignment, the Jungle Owlet sample had an average depth of 30X and the Spotted Owlet 26.5X. Genome coverage, depth filters, and average depth are expected to impact the outcome of PSMC and for reliable results, 75% of genome coverage with a 10X depth filter is recommended (Nadachowska-Brzyska et al., 2016). The genome coverage for both samples decreased with quality filtering and removal of non-autosomal scaffolds (Supplementary Figure S1) but was within the recommended limit (Nadachowska-Brzyska et al., 2016).
The parameters chosen for PSMC resulted in a time-span covering the LIG, the LGM, and the MDH i.e., the period of interest. Altering the mutation rate and the generation time based on other studies on owls and other birds (Nadachowska-Brzyska et al., 2013; Fujito, 2020) had predictable effects on the PSMC curve, as seen in other studies (Nadachowska-Brzyska et al., 2015; Vijay et al., 2018), but did not alter the shape (Supplementary Figure S2).
The two species show contrasting trends of population size (Ne) change over the past 500,000 years (Figure 1). The Ne of the Jungle Owlet rose gradually from approximately 14,500 during the LIG (120,000 YA) to about 78,000 during the LGM (22,000 YA) and eventually to about 160,000 at the MDH (6,000 YA). In contrast, the Spotted Owlet had the highest Ne, about 293,000, around the LGM. It rose from about 33,000 around the LIG and again fell after the LGM to about 45,000 around the MDH. It must be kept in mind that PSMC results are less robust on recent timescales (Li and Durbin, 2012; Patton et al., 2019) and this affects the Ne estimates around the Mid-Holocene. However, the population trends leading up from the LGM to the MDH still indicate divergent trajectories for the Jungle and Spotted Owlets. PSMC plots for the two owlets without scaling are shown in Supplementary Figure S3 and show the same contrasting pattern.
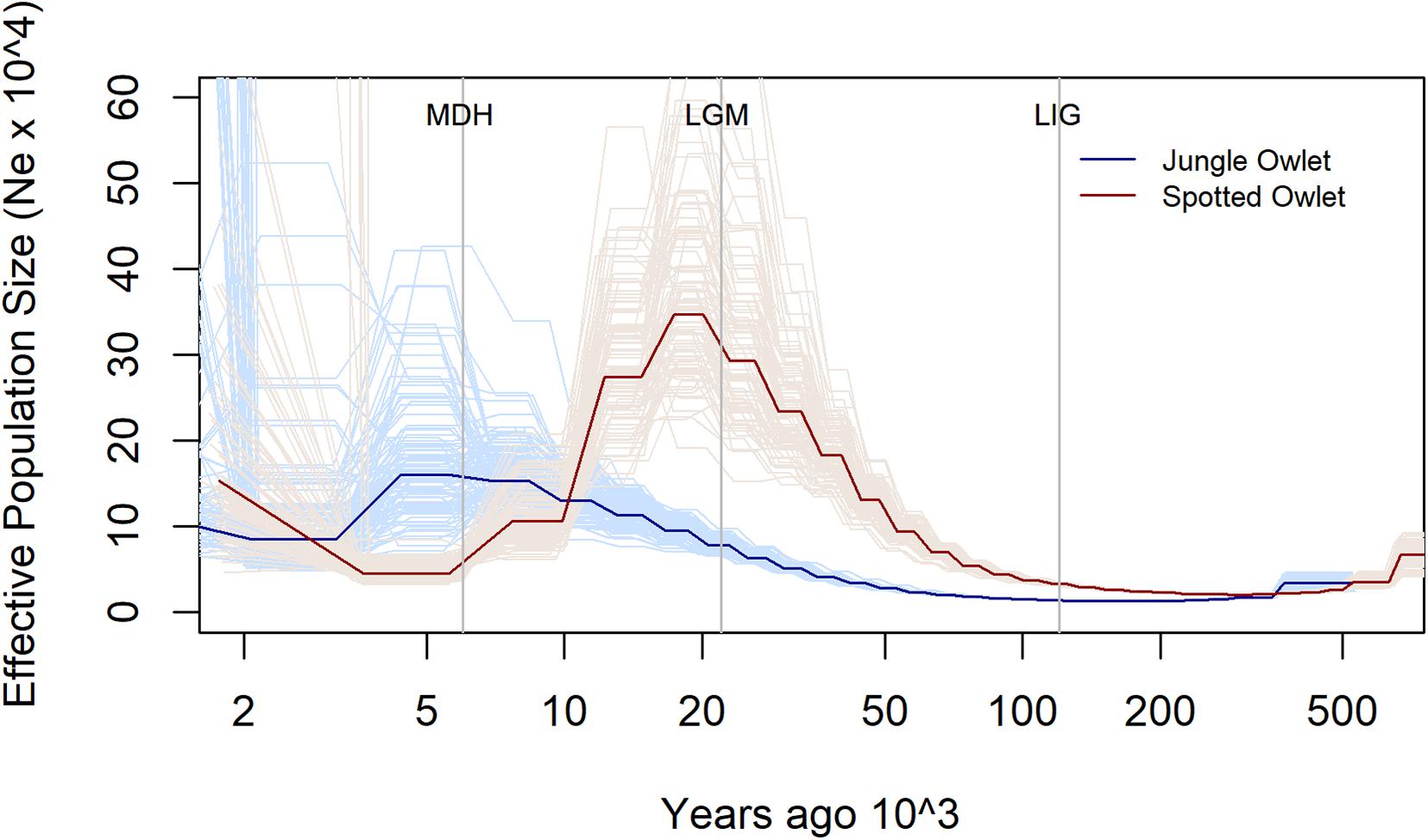
Figure 1. Estimated change in effective population size (Ne) for the Jungle (Glaucidium radiatum, dark blue) and Spotted (Athene brama, dark red) Owlets over the past 500,000 years. The pale lines denote estimates from the 100 bootstrap replicates for each species. The vertical lines denote significant climatic transitions in the past—the Last Interglacial (LIG—120,000 YBP), the Last Glacial Maximum (LGM—22,000 YBP), and the Mid-Holocene (MDH—6,000 YBP). To scale the plot, a mutation rate of 4 × 10–9 per site per year and a generation time of 2 years were used.
Discussion
In this study, we elucidate population size trends for the Jungle Owlet and Spotted Owlet over the major climatic events of the past—the LIG—beginning of the Last Glacial Period, the LGM, and the MDH. We find that both species show expansion from the LIG to the LGM, although the Jungle Owlet has a much greater degree of expansion. While the Jungle Owlet continues to expand into the MDH, the Spotted Owlet shows a declining trend after the LGM. The direction of these population trends is broadly consistent with predictions from the species distribution models from a previous study (Koparde et al., 2019), suggesting that their distinct niches had a significant impact on their population history. We note that we have sampled one individual of each species and our data may not be representative of trends across the range of these species. Therefore, we restrict our inferences to the sampled Western Ghats populations of these widely distributed species.
Temporal Trends in Ne Over the Late Quaternary for the Two Owlet Species
The LIG was a warm and wet period in India (Prabhu et al., 2004), with a strong southwest monsoon. Both samples show reduced Ne during this period, a pattern observed in multiple species from East-Asia (Dong et al., 2017). The LGM was characterized by a cold and dry climate, increased aridification, and reduced precipitation (Prabhu et al., 2004). In the Indian peninsula, the rain forests were replaced by grassland vegetation (Sukumar et al., 1995). These shifts are likely to have been favorable for the Spotted Owlet, which shows a considerable increase in population size toward the LGM. The Jungle Owlet also shows an increasing trend, albeit to a much lower degree, possibly due to the species’ preference for moist—dry forest and scrublands in potential refugia. Following this, the transition toward the Holocene climate resulted in a rise in temperature and precipitation. During this time, much of peninsular India shifted to wetter, forest habitats (Prabhu et al., 2004). The Spotted Owlet shows a decline in this period, consistent with the prediction of retraction of grasslands and open habitats, while the Jungle Owlet continues to show an upward trend.
One major concern with the PSMC model is the presence of population structure which has a confounding effect on the inference of population size changes (Mazet et al., 2015, 2016). For this reason, it is also possible that the population decline seen in the Spotted Owlet after the LGM may also be due to changes in population structure, and this needs to be tested with more data. Disentangling the effects of population structure and demography remains challenging, particularly with sequentially Markovian Coalescent methods (Mather et al., 2020). Another technical constraint is the absence of a reference genome assembly of the study species, which would have allowed better mapping of reads. Nevertheless, sampling more individuals and inclusion of more populations from across the range, while not logistically feasible for this study, may provide more conclusive results in the future.
The Influence of the Last Glacial Period on Species’ History in the Tropics
The Last Glacial Period has been a time of major population size changes for many avian species (Nadachowska-Brzyska et al., 2015). However, the direction and extent of the impact may differ across species within the same region (Cabanne et al., 2016) and within species across different regions (Nadachowska-Brzyska et al., 2016). Moreover, the impacts of the Last Glacial Period were variable within the tropics. While African forests retracted into refugial fragments in favor of grassland expansion during the dry, cool LGM period, Amazonian forests may have been relatively stable, even though the forest composition itself was altered (Prentice et al., 2000; Lessa et al., 2003; Malhi and Phillips, 2004; Bush and de Oliveira, 2006; Piñeiro et al., 2017). A study on 11 East-Asian birds, on the other hand, shows a greater impact of the LIG, where several species show range contraction, as opposed to the glacial period, which is a period of expansion (Dong et al., 2017).
In the Indian subcontinent, changes in diversification rates and population history have been shown to coincide with paleoclimatic shifts (Robin et al., 2010; Agarwal and Ramakrishnan, 2017). A high-altitude forest specialist bird, Sholicola albiventris, shows post-glacial population expansion, aligning well with the idea of forest fragmentation during the LGM (Robin et al., 2010). On the other hand, the Indian peafowl Pavo cristatus—a generalist (occupying moist-dry deciduous forests, to open forests and woodlands, and cultivated areas and orchards), shows a stable population size throughout the Last Glacial Period, despite earlier bottlenecks (Jaiswal et al., 2018). The Spotted Owlet in our study, in line with observations for many other species, shows a cyclic expansion and contraction of population size around the LGM, whereas the Jungle Owlet does not. Such differences could result from differences in the extent of habitat change and the strength of the habitat association. Moreover, our Jungle Owlet sample is from the more widely distributed subspecies Glaucidium radiatum radiatum, collected from central India. The subspecies from Southern India—Glaucidium radiatum malabaricum, known to be associated with more moist forests (Ali and Ripley, 1983), may show a population history more consistent with other forest-associated species.
Ne Trends in Understanding Long-Term Population History
The demographic reconstruction of population history of the two Owlet species appears to be broadly consistent with predictions from previously generated climatic niche models (Koparde et al., 2019) in terms of the direction of change. PSMC-based demographic inference coupled with Climatic niche models can be an effective means of understanding the population history of species (Kozma et al., 2018; Mays et al., 2018). Apart from indicating past population size trends, they are an important indicator of species vulnerability. Long-term declines have been observed in species that are now of conservation concern (Nadachowska-Brzyska et al., 2015) and are often considered to indicate reduced genetic variation and increased genetic load (Xue et al., 2015). While inferences from Ne curves alone are valuable, incorporating niche models allows correlation with past climate and vegetation. Moreover, population trends may not be a simple reflection of past climatic and vegetation changes. Other factors such as the dispersal ability of a species or the rate of change of suitable habitat may interact with the prevailing climatic and vegetation conditions to modify the genetic consequences of range contraction (Arenas et al., 2012). Therefore, genomic data can be highly valuable when combined with climatic niche models to test predictions about the impact of past climatic changes on species history.
The two owlet species in our study show evidence for differential impacts of Quaternary climatic events, likely driven by their differing ecological niches, despite the phylogenetic similarity. Given our limited sampling, we highlight the need for a larger investigation of the impacts of past climate on South Asian taxa to understand the community and species-specific impacts of past climate change.
Data Availability Statement
The raw illumina sequences for the two species have been submitted to NCBI and are available from the bioproject PRJNA665335.
Ethics Statement
Permit requests for this animal study were reviewed, approved, and issued by the State Forest Departments of Maharashtra and Kerala.
Author Contributions
SM, NV, and VVR: conceptualization. SM and VVR: resources. SG and KLV: lab work. MN: analysis. NV and VVR: supervision. MN, KLV, and VVR: writing. MN, KLV, SM, NV, VVR, and RJ: review and editing. VVR, SM, and RJ: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by MoEFCC grant—File Number J.22012/61/2009-CS(W), dated 18th June 2019 to SM, RJ, and VVR; and a SERB ECR grant ECR/2016/001065 to VVR.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the State Forest Departments of Maharashtra and Kerala for permits (permit letter numbers Desk-22(8)WL/CR-59(17-18)631/2019-20 and WL10-16346/2017, respectively); the Scientific Computing facility at IISER Tirupati and members of IT Department for HPC access and help with trouble-shooting; The Director, along with the Library, Administrative and Finance staff of SACON for logistic support; Naman Goyal, Ashwin Varudkar, and C. K. Vishnudas for help with sample collection. We are also grateful to the Ecology Evolution labs at IISER Tirupati for feedback on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.608339/full#supplementary-material
Footnotes
- ^ http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- ^ http://broadinstitute.github.io/picard/
- ^ https://github.com/meghananatesh/JO
References
Agarwal, I., and Ramakrishnan, U. (2017). A phylogeny of open-habitat lizards (squamata: lacertidae: Ophisops) supports the antiquity of Indian grassy biomes. J. Biogeogr. 44, 2021–2032. doi: 10.1111/jbi.12999
Ali, S., and Ripley, D. (1983). Handbook of the Birds of India and Pakistan. Volume 3: Stone-curlews to Owls, 2nd Edn. New Delhi: Oxford University Press.
Arenas, M., Ray, N., Currat, M., and Excoffier, L. (2012). Consequences of range contractions and range shifts on molecular diversity. Mol. Biol. Evol. 29, 207–218. doi: 10.1093/molbev/msr187
Batalha-Filho, H., Cabanne, G. S., and Miyaki, C. Y. (2012). Phylogeography of an Atlantic forest passerine reveals demographic stability through the last glacial maximum. Mol. Phylogenet. Evol. 65, 892–902. doi: 10.1016/j.ympev.2012.08.010
Beyer, R. M., Krapp, M., and Manica, A. (2020). High-Resolution terrestrial climate, bioclimate and vegetation for the last 120,000 years. Sci. Data 7:236. doi: 10.1038/s41597-020-0552-1
Bush, M. B., and de Oliveira, P. E. (2006). The rise and fall of the Refugial Hypothesis of Amazonian speciation: a paleoecological perspective. Biota Neotrop. 6. doi: 10.1590/S1676-06032006000100002
Cabanne, G. S., Calderón, L., Arias, N. T., Flores, P., Pessoa, R., D’Horta, F. M., et al. (2016). Effects of pleistocene climate changes on species ranges and evolutionary processes in the neotropical Atlantic forest. Biol. J. Linn. Soc. 119, 856–872. doi: 10.1111/bij.12844
Cabanne, G. S., Santos, F. R., and Miyaki, C. Y. (2007). Phylogeography of Xiphorhynchus fuscus (Passeriformes, Dendrocolaptidae): vicariance and recent demographic expansion in Southern Atlantic forest. Biol. J. Linn. Soc. 91, 73–84. doi: 10.1111/j.1095-8312.2007.00775.x
Dong, F., Hung, C. M., Li, X. L., Gao, J. Y., Zhang, Q., Wu, F., et al. (2017). Ice age unfrozen: severe effect of the last interglacial, not glacial, climate change on East Asian avifauna. BMC Evol. Biol. 17:244. doi: 10.1186/s12862-017-1100-2
Dynesius, M., and Jansson, R. (2000). Evolutionary consequences of changes in species’ geographical distributions driven by milankovitch climate oscillations. Proc. Natl. Acad. Sci. U.S.A. 97, 9115–9120. doi: 10.1073/pnas.97.16.9115
Feng, S., Fang, Q., Barnett, R., Li, C., Han, S., Kuhlwilm, M., et al. (2019). The genomic footprints of the fall and recovery of the crested ibis. Curr. Biol. 29, 340–349.e7. doi: 10.1016/j.cub.2018.12.008
Fuchs, J., Alstrom, P., Yosef, R., and Olsson, U. (2019). Miocene diversification of an open-habitat predatorial passerine radiation, the shrikes (aves: passeriformes: laniidae). Zool. Scr. 48, 571–588. doi: 10.1111/zsc.12363
Fujito, N. T., Hanna, Z. R., Levy-Sakin, M., Bowie, R. C. K., Kwok, P.-Y., Dumbacher, J. P., et al. (2020). Genomic variation and population histories of spotted (Strix occidentalis) and barred (S. varia) owls. bioRxiv 2020.02.18.954685. doi: 10.1101/2020.02.18.954685
Hewitt, G. (2000). The genetic legacy of the quaternary ice ages. Nature 405, 907–913. doi: 10.1038/35016000
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the quaternary. Philos. Trans. R. Soc. B Biol. Sci. 359, 183–195. doi: 10.1098/rstb.2003.1388
Hofreiter, M., and Stewart, J. (2009). Ecological change, range fluctuations and population dynamics during the pleistocene. Curr. Biol. 19, R584–R594. doi: 10.1016/j.cub.2009.06.030
Holt, D. W., Berkley, R., Deppe, C., Enríquez, P. L., Petersen, J. L., Rangel Salazar, J. L., et al. (2020a). “Jungle owlet (Glaucidium radiatum), version 1.0,” in Birds of the World, eds J. del Hoyo, A. Elliott, J. Sargatal, D. A. Christie, and E. de Juana (Ithaca, NY: Cornell Lab of Ornithology), doi: 10.2173/bow.junowl1.01
Holt, D. W., Berkley, R., Deppe, C., Enríquez, P. L., Petersen, J. L., Rangel Salazar, J. L., et al. (2020b). “Spotted owlet (Athene brama), version 1.0,” in Birds of the World, eds J. del Hoyo, A. Elliott, J. Sargatal, D. A. Christie, and E. de Juana (Ithaca, NY: Cornell Lab of Ornithology), doi: 10.2173/bow.spoowl1.01
Hung, C. M., Shaner, P. J. L., Zink, R. M., Liu, W. C., Chu, T. C., Huang, W. S., et al. (2014). Drastic population fluctuations explain the rapid extinction of the passenger pigeon. Proc. Natl. Acad. Sci. U.S.A. 111, 10636–10641. doi: 10.1073/pnas.1401526111
Jaiswal, S. K., Gupta, A., Saxena, R., Prasoodanan, V. P. K., Sharma, A. K., Mittal, P., et al. (2018). Genome sequence of peacock reveals the peculiar case of a glittering bird. Front. Genet. 9:392. doi: 10.3389/fgene.2018.00392
Koparde, P., Mehta, P., Mukherjee, S., and Robin, V. V. (2019). Quaternary climatic fluctuations and resulting climatically suitable areas for Eurasian owlets. Ecol. Evol. 9, 4864–4874. doi: 10.1002/ece3.5086
Kozma, R., Lillie, M., Benito, B. M., Svenning, J. C., and Höglund, J. (2018). Past and potential future population dynamics of three grouse species using ecological and whole genome coalescent modeling. Ecol. Evol. 8, 6671–6681. doi: 10.1002/ece3.4163
Kozma, R., Melsted, P., Magnússon, K. P., and Höglund, J. (2016). Looking into the past – the reaction of three grouse species to climate change over the last million years using whole genome sequences. Mol. Ecol. 25, 570–580. doi: 10.1111/mec.13496
Lessa, E. P., Cook, J. A., and Patton, J. L. (2003). Genetic footprints of demographic expansion in North America, but not Amazonia, during the late quaternary. Proc. Natl. Acad. Sci. U.S.A. 100, 10331–10334. doi: 10.1073/pnas.1730921100
Li, H. (2011). A statistical framework for snp calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv [preprint] 1–3. Available online at: https://arxiv.org/abs/1303.3997 (accessed August 1, 2020).
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., and Durbin, R. (2012). Inference of human population history from whole genome sequence of a single individual. Nature 475, 493–496. doi: 10.1038/nature10231.Inference
Lucena-Perez, M., Marmesat, E., Kleinman-Ruiz, D., Martínez-Cruz, B., Węcek, K., Saveljev, A. P., et al. (2020). Genomic patterns in the widespread eurasian lynx shaped by late quaternary climatic fluctuations and anthropogenic impacts. Mol. Ecol. 29, 812–828. doi: 10.1111/mec.15366
Malhi, Y., and Phillips, O. L. (2004). Tropical forests and global atmospheric change: a synthesis. Philos. Trans. R. Soc. B Biol. Sci. 359, 549–555. doi: 10.1098/rstb.2003.1449
Mather, N., Traves, S. M., and Ho, S. Y. W. (2020). A practical introduction to sequentially Markovian coalescent methods for estimating demographic history from genomic data. Ecol. Evol. 10, 579–589. doi: 10.1002/ece3.5888
Mays, H. L., Hung, C. M., Shaner, P. J., Denvir, J., Justice, M., Yang, S. F., et al. (2018). Genomic analysis of demographic history and ecological niche modeling in the endangered Sumatran rhinoceros Dicerorhinus sumatrensis. Curr. Biol. 28, 70–76.e4. doi: 10.1016/j.cub.2017.11.021
Mazet, O., Rodríguez, W., and Chikhi, L. (2015). Demographic inference using genetic data from a single individual: separating population size variation from population structure. Theor. Popul. Biol. 104, 46–58. doi: 10.1016/j.tpb.2015.06.003
Mazet, O., Rodríguez, W., Grusea, S., Boitard, S., and Chikhi, L. (2016). On the importance of being structured: instantaneous coalescence rates and human evolution-lessons for ancestral population size inference? Heredity 116, 362–371. doi: 10.1038/hdy.2015.104
McKenna, A., Hanna, M., Banks, E., Sivachenko, A. Y., Cibulskis, K., Kernytsky, A. M., et al. (2010). The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110.20
Mehta, P., Kulkarni, J., Talmale, S., and Chandarana, R. (2018). Diets of sympatric forest owlets, spotted owlets, and jungle owlets in East Kalibhit forests, Madhya Pradesh, India. J. Raptor Res. 52, 338–348. doi: 10.3356/JRR-17-00002.1
Mueller, J. C., Kuhl, H., Boerno, S., Tella, J. L., Carrete, M., and Kempenaers, B. (2018). Evolution of genomic variation in the burrowing owl in response to recent colonization of Urban areas. Proc. R. Soc. B Biol. Sci. 285:20180206. doi: 10.1098/rspb.2018.0206
Nadachowska-Brzyska, K., Burri, R., Olason, P. I., Kawakami, T., Smeds, L., and Ellegren, H. (2013). Demographic divergence history of pied flycatcher and collared flycatcher inferred from whole-genome re-sequencing data. PLoS Genet. 9:e1003942. doi: 10.1371/journal.pgen.1003942
Nadachowska-Brzyska, K., Burri, R., Smeds, L., and Ellegren, H. (2016). PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol. Ecol. 25, 1058–1072. doi: 10.1111/mec.13540
Nadachowska-Brzyska, K., Li, C., Smeds, L., Zhang, G., and Ellegren, H. (2015). Temporal dynamics of avian populations during pleistocene revealed by whole-genome sequences. Curr. Biol. 25, 1375–1380. doi: 10.1016/j.cub.2015.03.047
Pande, S., Pawashe, A., Mahajan, M. N., Joglekar, C., and Mahabal, A. (2007). Effect of food and habitat on breeding success in spotted owlets (Athene brama) nesting in villages and rural landscapes in India. J. Raptor Res. 41, 26–34. doi: 10.3356/0892-1016(2007)41[26:eofaho]2.0.co;2
Patton, A. H., Margres, M. J., Stahlke, A. R., Hendricks, S., Lewallen, K., Hamede, R. K., et al. (2019). Contemporary demographic reconstruction methods are robust to genome assembly quality: a case study in Tasmanian devils. Mol. Biol. Evol. 36, 2906–2921. doi: 10.1093/molbev/msz191
Piñeiro, R., Dauby, G., Kaymak, E., and Hardy, O. J. (2017). Pleistocene. population expansions of shade-tolerant trees indicate fragmentation of the african rainforest during the ice ages. Proc. R. Soc. B Biol. Sci. 284:20171800. doi: 10.1098/rspb.2017.1800
Prabhu, C. N., Shankar, R., Anupama, K., Taieb, M., Bonnefille, R., Vidal, L., et al. (2004). A 200-Ka pollen and oxygen-isotopic record from two sediment cores from the Eastern Arabian sea. Palaeogeogr. Palaeoclimatol. Palaeoecol. 214, 309–321. doi: 10.1016/j.palaeo.2004.07.027
Prentice, I. C., and Jolly, D., and Biome 6000 Participants. (2000). Mid-Holocene and glacial-maximum vegetation geography of the northern continents and Africa. J. Biogeogr. 27, 507–519. doi: 10.1046/j.1365-2699.2000.00425.x
Qian, H., and Ricklefs, R. E. (2001). Palaeovegetation diversity of temperate plants in East Asia. Nature 413, 129–130. doi: 10.1038/35093169
Ray, N., and Adams, J. M. (2001). A GIS-based vegetation map of the world at the last glacial maximum (25,000-15,000 BP). Internet Archaeol. 11, 1–44.
Reddy, S. (2014). What’s missing from Avian global diversification analyses? Mol. Phylogenet. Evol. 77, 159–165. doi: 10.1016/j.ympev.2014.04.023
Robin, V. V., Sinha, A., and Ramakrishnan, U. (2010). Ancient geographical gaps and paleo-climate shape the phylogeography of an endemic bird in the sky Islands of Southern India. PLoS One 5:e13321. doi: 10.1371/journal.pone.0013321
Rocha, D. G. d., and Kaefer, I. L. (2019). What has become of the Refugia hypothesis to explain biological diversity in Amazonia? Ecol. Evol. 9, 4302–4309. doi: 10.1002/ece3.5051
Schneider, C., and Moritz, C. (1999). Rainforest Refugia and evolution in Australia’s wet tropics. Proc. R. Soc. B Biol. Sci. 266, 191–196. doi: 10.1098/rspb.1999.0621
Song, K., Gao, B., Halvarsson, P., Fang, Y., Jiang, Y.-X., Sun, Y.-H., et al. (2020). Genomic analysis of demographic history and ecological niche modeling in the endangered Chinese grouse Tetrastes Sewerzowi. BMC Genom. 21:581. doi: 10.21203/rs.2.17686/v1
Sukumar, R., Suresh, H. S., and Ramesh, R. (1995). Climate change and its impact on tropical montane ecosystems in Southern India. J. Biogeogr. 22, 533–536. doi: 10.2307/2845951
Vijay, N., Park, C., Oh, J., Jin, S., Kern, E., Kim, H. W., et al. (2018). Population genomic analysis reveals contrasting demographic changes of two closely related Dolphin species in the last glacial. Mol. Biol. Evol. 35, 2026–2033. doi: 10.1093/molbev/msy108
Wurster, C. M., Bird, M. I., Bull, I. D., Creed, F., Bryant, C., Dungait, J. A. J., et al. (2010). Forest contraction in north equatorial Southeast Asia during the last glacial period. Proc. Natl. Acad. Sci. U.S.A. 107, 15508–15511. doi: 10.1073/pnas.1005507107
Xue, Y., Prado-martinez, J., Sudmant, P. H., Narasimhan, V., Ayub, Q., Szpak, M., et al. (2015). Mountain Gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348, 242–245. doi: 10.1126/science.aaa3952
Keywords: PSMC, population genomics, South Asia/India, climatic niche models, Owlets, last glacia maximum
Citation: Natesh M, Vinay KL, Ghosh S, Jayapal R, Mukherjee S, Vijay N and Robin VV (2020) Contrasting Trends of Population Size Change for Two Eurasian Owlet Species—Athene brama and Glaucidium radiatum From South Asia Over the Late Quaternary. Front. Ecol. Evol. 8:608339. doi: 10.3389/fevo.2020.608339
Received: 20 September 2020; Accepted: 30 November 2020;
Published: 22 December 2020.
Edited by:
Shan Huang, Senckenberg Biodiversity and Climate Research Centre, GermanyReviewed by:
Baocheng Guo, Chinese Academy of Sciences (CAS), ChinaReuven Yosef, Ben-Gurion University of the Negev, Israel
Copyright © 2020 Natesh, Vinay, Ghosh, Jayapal, Mukherjee, Vijay and Robin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meghana Natesh, meghana@iisertirupati.ac.in; meg.nat86@gmail.com; V. V. Robin, robin@iisertirupati.ac.in; orcid.org/0000-0003-3109-5498
†These authors have contributed equally to this work
 Meghana Natesh
Meghana Natesh K. L. Vinay
K. L. Vinay Samriddha Ghosh2
Samriddha Ghosh2  Rajah Jayapal
Rajah Jayapal Shomita Mukherjee
Shomita Mukherjee Nagarjun Vijay
Nagarjun Vijay V. V. Robin
V. V. Robin