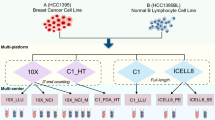
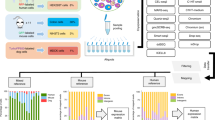
Abstract
Comparing diverse single-cell RNA sequencing (scRNA-seq) datasets generated by different technologies and in different laboratories remains a major challenge. Here we address the need for guidance in choosing algorithms leading to accurate biological interpretations of varied data types acquired with different platforms. Using two well-characterized cellular reference samples (breast cancer cells and B cells), captured either separately or in mixtures, we compared different scRNA-seq platforms and several preprocessing, normalization and batch-effect correction methods at multiple centers. Although preprocessing and normalization contributed to variability in gene detection and cell classification, batch-effect correction was by far the most important factor in correctly classifying the cells. Moreover, scRNA-seq dataset characteristics (for example, sample and cellular heterogeneity and platform used) were critical in determining the optimal bioinformatic method. However, reproducibility across centers and platforms was high when appropriate bioinformatic methods were applied. Our findings offer practical guidance for optimizing platform and software selection when designing an scRNA-seq study.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets generated and analyzed in the current study are available in the SRA repository under accession code no. PRJNA504037. The data from Tian et al. are available at GEO accession code GSE118767. The data from Hie et al. are available at the following URL: http://scanorama.csail.mit.edu/data.tar.gz.
Code availability
We used many algorithms and code sets for batch correction that were previously published. All of our code is provided in GitHub and Code Ocean at the following links: https://github.com/oxwang/fda_scRNA-seq and https://codeocean.com/capsule/0497386 or https://doi.org/10.24433/CO.1559060.v1.
References
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Gierahn, T. M. et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017).
Liu, T., Wu, H., Wu, S. & Wang, C. Single-cell sequencing technologies for cardiac stem cell studies. Stem Cells Dev. 26, 1540–1551 (2017).
Wu, H., Wang, C. & Wu, S. Single-cell sequencing for drug discovery and drug development. Curr. Top. Med. Chem. 17, 1769–1777 (2017).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Hie, B., Bryson, B. & Berger, B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. 37, 685–691 (2019).
Polanski, K. et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 36, 964–965 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019).
Ziegenhain, C. et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643 (2017).
Zhang, X. et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell 73, 130–142 (2019).
Svensson, V. et al. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14, 381–387 (2017).
Mereu, E. et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 38, 747–755 (2020).
Tian, L. et al. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat. Methods 16, 479–487 (2019).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Gazdar, A. F. et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int. J. Cancer 78, 766–774 (1998).
Xiao, W. et al. Towards best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat. Biotechnol. (in the press).
Zhang, J., Spath, S. S., Marjani, S. L., Zhang, W. & Pan, X. Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment. Precis. Clin. Med. 1, 29–48 (2018).
Chen, X. et al. A multi-center cross-platform single-cell RNA sequencing reference dataset. Preprint at bioRxiv https://doi.org/10.1101/2020.09.20.305474 (2020).
Zhang, M. J., Ntranos, V. & Tse, D. Determining sequencing depth in a single-cell RNA-seq experiment. Nat. Commun. 11, 774 (2020).
Li, B. et al. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods 17, 793–798 (2020).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017).
Parekh, S., Ziegenhain, C., Vieth, B., Enard, W. & Hellmann, I. zUMIs—a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7, giy059 (2018).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. J. B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Hicks, S. C., Townes, F. W., Teng, M. & Irizarry, R. A. Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics. 19, 562–578 (2017).
Risso, D., Ngai, J., Speed, T. P. & Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Lun, A. T., Bach, K. & Marioni, J. C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 17, 75 (2016).
Bacher, R. et al. SCnorm: robust normalization of single-cell RNA-seq data. Nat. Methods 14, 584–586 (2017).
Yip, S. H., Wang, P., Kocher, J.-P. A., Sham, P. C. & Wang, J. Linnorm: improved statistical analysis for single cell RNA-seq expression data. Nucleic Acids Res. 45, e179 (2017).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Yip, S. H., Sham, P. C. & Wang, J. Evaluation of tools for highly variable gene discovery from single-cell RNA-seq data. Brief Bioinform. 20, 1583–1589 (2018).
Buettner, F. et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 33, 155–160 (2015).
Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94 (2018).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Buttner, M., Miao, Z., Wolf, F. A., Teichmann, S. A. & Theis, F. J. A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods 16, 43–49 (2019).
Kaminski, D. A., Wei, C., Qian, Y., Rosenberg, A. F. & Sanz, I. Advances in human B cell phenotypic profiling. Front. Immunol. 3, 302 (2012).
Starlets, D. et al. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood 107, 4807–4816 (2006).
Zook, J. M. et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 32, 246–251 (2014).
Alles, J. et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 15, 44 (2017).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Krueger, F. Trim Galore! http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (2015).
Cole, M. B. et al. Performance assessment and selection of normalization procedures for single-cell RNA-seq. Cell Syst. 8, 315–328 (2019).
Qiu, X. et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Acknowledgements
We thank D. Ho of the LLU Center for Genomics for her great administrative support, particularly in coordinating the weekly Zoom conference calls and assistance in preparing meeting minutes for the SEQC-2 single-cell sequencing project. We thank ATCC, and particularly L. Kerrigan, for providing the two cell lines, that is, HCC1395 and HCC1395BL, for our study. We thank W. Jones at EA Genomics, Q2 Solutions for critical review and helpful comments. We thank Z. Chen at LLU and J. Shetty at the NCI for technical assistance in performing sequencing, J. Bettridge at the NCI for technical assistance in 10x Genomics scRNA-seq library preparation, V. Furtak at the FDA for library preparation and W. Wu at the FDA/CBER Core Facility for Illumina sequencing. We also thank S. Anandakrishnan of Takara Bio USA, Inc. for technical assistance with TBU ICELL8 single-cell capture and library preparation. The genomic work carried out at the LLU Center for Genomics was funded in part by the National Institutes of Health (NIH) grant S10OD019960 (C.W.), the Ardmore Institute of Health grant 2150141 (C.W.) and C.A. Sims’ gift to LLU Center for Genomics.
Author information
Authors and Affiliations
Contributions
C.W. and W.X. conceived and designed the study. C.W. managed the project and directed bioinformatic data analyses. C.W. drafted the manuscript and annotated all the results. M.M. Jr. and A.F. helped edit the manuscript. W.C., B.T., M.M., P.K., M.M. Jr., A.F. and A.M. performed single-cell culturing, single-cell captures and scRNA-seq library synthesis and sequencing. X.C., Z.Y., Y.Z., X.X., V.C., Y.B., B.E., W.X., U.A.M., J.L., J.-L.L. and C.W. performed bioinformatic data analyses. W.C., X.C., Z.Y., Y.Z., Y.B., X.X., V.C., M.M., A.M., M.M. Jr. and J.-L.L. prepared the methods for the manuscript. Z.Y. prepared all figures; W.C., C.W. and H.C. prepared the tables. C.W., M.M. Jr., W.C., A.F. and W.X. revised the manuscript. All authors reviewed the manuscript. C.W. finalized and submitted the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.F. and A.M. are employees of Takara Bio USA, Inc., and B.E. and U.A.M. were employees of Digicon Corporation. All other authors claim no conflicts of interest. The views presented in this article do not necessarily reflect current or future opinion or policy of the US Food and Drug Administration. Any mention of commercial products is for clarification and not intended as an endorsement.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 An overview of the number of genes detected in each cell across all datasets.
The violin plot shows the number of genes detected in each cell across 20 scRNA-seq datasets. The plot was generated using Seurat (version 3.1). Each dot represents a single cell. The violin shapes summarize the data distributions, which are colored in the background to signify each of the 20 different scRNA seq datasets. Each scRNA-seq dataset is plotted on the X-axis; the Y-axis shows the corresponding number of genes detected in a cell (nGene) for that dataset. The average number of genes detected in each cell was about 4000 and most of the cells had 2500–7500 genes, except for samples C1_LLU_A and C1_LLU_B. The 10x Genomics scRNA datasets were preprocessed using Cell Ranger version 3.1.
Extended Data Fig. 2 Regressing mitochondrial genes and normalizing UMI did not remove batch effects.
Five different batches of scRNA-seq data (10X_LLU_A, 10X_LLU_B, 10X_NCI_A, 10X_NCI_B, and 10X_NCI_Mix5) generated at two sites (LLU and NCI) are shown either as t-SNE plots (panels a-d) or as UMAPs (panels e-h). (a) logNormalized, scaled data with no regression; (b) logNormalized, scaled data filtered with mitochondrial (Mito) gene regression >5% and UMI normalization by Seurat v3; (c) sctransform with no regression; (d) sctransform with mitochondrial gene regression and UMI normalization; (e) logNormalized, scaled data with no regression; (f) scaled data with mitochondrial gene regression and UMI normalization; (g) sctransform with no regression; and (h) sctransform with mitochondrial gene regression and UMI normalization.
Extended Data Fig. 3 UMAP showing batch-effect correction by mixability and clusterability using scRNA-seq datasets in four different sample scenarios.
Batch-effect corrections were performed for the following four scenarios: (a) Scenario 1, where all 20 scRNA-seq datasets were combined, including mixed and non-mixed, with large proportions of two dissimilar types of cells (sample A, breast cancer cell line HCC1395 and sample B, B-lymphocyte line HCC1395BL); Datasets from 10x were down-sampled to 1200 cells per dataset. (b) Scenario 2, where five datasets (10X_LLU_A, 10X_NCI_A, C1_FDA_HT_A, C1_LLU_A, and ICELL8_SE_A) from the breast cancer cells (sample A, HCC1395) were generated separately at four centers (LLU, NCI, FDA, and TBU) on four platforms (10x, Fluidigm C1, Fluidigm C1_HT, and TBU ICELL8); (c) Scenario 3, where five datasets (10X_LLU_B, 10X_NCI_B, C1_FDA_HT_B, C1_LLU_B, and ICELL8_SE_B) from B-lymphocytes (sample B, HCC1395BL) were generated separately at four centers (LLU, NCI, FDA, and TBU) on four platforms (10x, Fluidigm C1, Fluidigm C1_HT, and TBU ICELL8); and (d) Scenario 4, where four datasets (10X_LLU_Mix10, 10X_NCI_M_Mix5, 10X_NCI_M_Mix5_F, and 10X_NCI_M_Mix5_F2) were generated from 5% or 10% of breast cancer cells (sample A, HCC1395), spiked into the B-lymphocytes (sample B, HCC1395BL), and analyzed with the 10x Genomics platform at two centers (LLU and NCI) in four different batches. Batch correction methods included Seurat v3.1, fastMNN (SeuratWrappers v0.1.0), Scanorama v1.4, BBKNN v1.3.5, Harmony v0.99.9, limma v3.40.4, and Combat (sva v3.32.1). The top 2000 highly variable genes (HVGs) of these datasets were used as the gene set for batch correction. All the 10x data were preprocessed using Cell Ranger version 3.1.
Extended Data Fig. 4 t-SNE plots and UMAPs showing batch-effect corrections by mixability and clusterability across four scRNA-seq platforms.
t-SNE plots and UMAPs showing the batch-effect corrections performed by seven methods using 20 scRNA-seq datasets across different platforms. Datasets from 10x were down-sampled to 1200 cells per dataset. *Note, for BBKNN, only UMAP was available and shown. The scRNA-seq datasets are colored to identify the four different platforms: 10x 3´ scRNA-seq platform (red), C1 3´ HT scRNA-seq platform (yellow), C1 full-length scRNA-seq platform (light blue), and ICELL8 full-length scRNA-seq platform (dark blue). Batch correction methods included: Seurat v3.1, fastMNN (SeuratWrappers v0.1.0), Scanorama v1.4, BBKNN v1.3.5, Harmony v0.99.9, limma v3.40.4, and Combat (sva v3.32.1). Scanorama failed to separate two cell types into discrete clusters when non-10x platforms were included in the analysis. The top 2000 HVGs across all datasets were used as the gene set for batch correction. All the 10x data were preprocessed using Cell Ranger version 3.1.
Extended Data Fig. 5 Batch-effect correction displayed by cell type identity.
Batch-effect corrections were performed for the following four scenarios: (a) Scenario 1, where all 20 scRNA-seq datasets were combined, including mixed and non-mixed, with large proportions of two dissimilar types of cells (sample A, breast cancer cell line HCC1395 and sample B, B-lymphocyte line HCC1395BL); Datasets from 10x were down-sampled to 1200 cells per dataset. (b) Scenario 2, where five datasets (10X_LLU_A, 10X_NCI_A, C1_FDA_HT_A, C1_LLU_A, and ICELL8_SE_A) from the breast cancer cells (sample A, HCC1395) were generated separately at four centers (LLU, NCI, FDA, and TBU) on four platforms (10x, Fluidigm C1, Fluidigm C1_HT, and TBU ICELL8); (c) Scenario 3, where five datasets (10X_LLU_B, 10X_NCI_B, C1_FDA_HT_B, C1_LLU_B, and ICELL8_SE_B) from B-lymphocytes (sample B, HCC1395BL) were generated separately at four centers (LLU, NCI, FDA, and TBU) on four platforms (10x, Fluidigm C1, Fluidigm C1_HT, and TBU ICELL8); and (d) Scenario 4, where four datasets (10X_LLU_Mix10, 10X_NCI_M_Mix5, 10X_NCI_M_Mix5_F, 10X_NCI_M_Mix5_F2) were generated from 5% or 10% of breast cancer cells (sample A, HCC1395) spiked into the B-lymphocytes (sample B, HCC1395BL) and analyzed with the 10x Genomics platform at two centers (LLU and NCI) in four different batches. *For BBKNN, only UMAPs were available and shown in (a–d); all others are t-SNE plots. The HCC1395 breast cancer cells (sample A) were labeled in red and the HCC1395BL B lymphocytes (sample B) were labeled in blue. Batch correction methods included Seurat v3.1, fastMNN (SeuratWrappers v0.1.0), Scanorama v1.4, BBKNN v1.3.5, Harmony v0.99.9, limma v3.40.4, and Combat (sva v3.32.1). The top 2000 HVGs were used as the gene set for batch correction. All the 10x data were preprocessed using Cell Ranger version 3.1.
Extended Data Fig. 6 Scanorama worked well for 10x Genomics scRNA-seq datasets regardless of the presence of shared cell types across batches.
(a) t-SNE plot and (b) UMAP showing batch-effect corrections using twelve 10x Genomics scRNA-seq datasets consisting of both mixed and non-mixed samples from two sites (LLU and NCI) in different batches after Scanorama (version 1.4.) batch correction. (c) t-SNE plot and (d) UMAP showing projections of batch-effect corrections using six 10x scRNA-seq datasets consisting of only non-mixed samples from two sites (LLU and NCI) in different batches after Scanorama (version 1.4.) batch correction. Different colors represent different datasets. All the datasets were down-sampled to 1200 cells per dataset. After the batch correction, cells from the same cell line type clustered together and mixed adequately within the same cell types. All the data were preprocessed using Cell Ranger version 3.1.
Extended Data Fig. 7 Batch-effect correction evaluating clusterability using 14 scRNA-seq datasets without spiked-in mixtures.
t-SNE plots and UMAPs showing batch-effect corrections performed by seven methods using 14 non-mixture scRNA-seq datasets across different platforms and sites. Six spiked-in mixture scRNA-seq datasets (10X_LLU_Mix10, 10X_NCI_Mix5, 10X_NCI_Mix5_F, 10X_NCI_M_Mix5, 10X_NCI_M_Mix5_F, and 10X_NCI_M_Mix5_F2) were removed from the 20 datasets in Scenario 1 for batch-effect correction evaluation. The fourteen non-mixture scRNA-seq datasets are from both breast cancer cells (10X_LLU_A, 10X_NCI_A, 10X_NCI_M_A, C1_FDA_HT_A, C1_LLU_A, ICELL8_SE_A, and ICELL8_PE_A) and B-lymphocytes (10X_LLU_B, 10X_NCI_B, 10X_NCI_M_B, C1_FDA_HT_B, C1_LLU_B, ICELL8_SE_B, and ICELL8_PE_B). Datasets from 10x were down-sampled to 1200 cells per dataset. *Note, for BBKNN, only UMAP was available and shown. Batch correction methods included Seurat v3.1, fastMNN (SeuratWrappers v0.1.0), Scanorama v1.4, BBKNN v1.3.5, Harmony v0.99.9, limma v3.40.4, and Combat (sva v3.32.1). All the 10x data were preprocessed using Cell Ranger version 3.1.
Extended Data Fig. 8 fastMNN batch-effect correction depends on the order of importing scRNA-seq data into the pipeline.
Panels (a-c) show results obtained using fastMNN when the spiked-in (mixed) datasets (that is, 10X_LLU_Mix10, 10X_NCI_Mix5, 10X_NCI_Mix5_F, 10X_NCI_M_Mix5, 10X_NCI_M_Mix5_F, and 10X_NCI_M_Mix5_F2) were imported into the pipeline before other non-mixed scRNA-seq datasets from the 20 scRNA-seq datasets of Scenario 1. (a) t-SNE vs. UMAP with color-coding by dataset; (b) tSNE vs. UMAP, colored by cell types (HCC1395, red; HCC1395BL, blue); and (c) A silhouette score = 0.52 showing that fastMNN correctly separated the two cell types into two clusters representing breast cancer cells and B lymphocytes. Panels (d-f) show results obtained using fastMNN when the non-mixed datasets were imported into the pipeline before the mixture datasets. (d) tSNE vs. UMAP with color-coding by datasets or (e) tSNE vs. UMAP colored by cell types; and (f) A low silhouette score of 0.22 showing that fastMNN had difficulty correctly separating the two cell types in this case. Batch-effect corrections were performed using fastMNN (SeuratWrappers v0.1.0) and silhouette width scores were calculated using the silhouette function from the R package cluster (v.2.0.8). Datasets from 10x were down-sampled to 1200 cells per dataset. The order of dataset input is shown on the top of the Figures (a, b, c or d, e, f).
Extended Data Fig. 9 Correlations of gene expression profiles across datasets.
Scatter plots displaying the gene expression profile correlations between each of seven scRNA-seq datasets (10X_LLU, 10X_NCI, 10X_NCI_M, C1_FDA, C1_LLU, ICELL8_SE, and ICELL8_PE) vs. their corresponding bulk RNA-seq dataset (BK_RNA-seq) for either (a) breast cancer cells or (b) B lymphocytes. The commonly detected transcripts [(log(CPM + 1) normalized] across all datasets were used (15,553 genes for breast cancer cells and 15,201 genes for B lymphocytes) to generate the scatter plots. Each dot represents each gene as a point in each scatterplot; x,y values represent the gene expression variation in a pair of compared datasets. The middle diagonal bar charts display the distribution of the most abundant or rare genes in each dataset and also provide the labels for the respective datasets. The Pearson correlation coefficient R between each of the datasets compared is shown to display the consistency of the different RNA-seq datasets.
Extended Data Fig. 10 Scanorama batch correction using 10x and non-10x scRNA-seq datasets from two different studies.
(a, un-corrected) UMAP of 10 datasets (10x: PBMCs 68 K, PBMCs 3 K, CD19 + B cells, CD14 + monocytes, CD4 + helper T cells, CD56 + NK cells, CD8 + cytotoxic T cells, CD4 + CD45RO + memory T cells, CD4 + CD25 + regulatory T cells; Drop-seq: PBMCs) out of 26 datasets from Hie et al.8 before batch correction by Scanorama. (b, corrected-based on dataset) UMAP of 10 different datasets shown in (a) from Hie et al. after batch correction by Scanorama, colored to identify the datasets. (c, corrected-based on platform) UMAP of 10 different datasets shown in (a) from Hie et al. colored to identify the two different platforms used (10x Genomics and Drop-seq); note poor results using Drop-seq. (d, un-corrected) UMAP of 8 datasets (breast cancer cells: C1_FDA_HT_A, C1_LLU_A, ICELL8_SE_A, and ICELL8_PE_A; and B lymphocytes: C1_FDA_HT_B, C1_LLU_B, ICELL8_SE_B, and ICELL8_PE_B) out of 20 datasets in our study analyzed using three different non-10x sequencing platforms before batch correction by Scanorama. (e, corrected-based on dataset) UMAP of 8 datasets shown in (d) after batch correction by Scanorama, colored to identify the datasets. Note lack of discrimination between different cell types. (f, corrected-based on platform) UMAP of 8 datasets shown in (d) after batch correction by Scanorama, colored to identify the platforms (C1_FDA_HT, blue; C1, purple; ICELL8, pink). The PBMC datasets were downloaded from http://scanorama.csail.mit.edu/data_light.tar.gz. Our eight datasets were preprocessed using the featureCounts pipeline and batch-effect correction was performed using Scanorama v1.4.
Supplementary information
Supplementary Information
Supplementary Figs. 1–14
Supplementary Data 1
DEGs based on DESeq analysis comparing breast cancer cells with B lymphocytes.
Rights and permissions
About this article
Cite this article
Chen, W., Zhao, Y., Chen, X. et al. A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples. Nat Biotechnol 39, 1103–1114 (2021). https://doi.org/10.1038/s41587-020-00748-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-00748-9
This article is cited by
-
Computational immunogenomic approaches to predict response to cancer immunotherapies
Nature Reviews Clinical Oncology (2024)
-
Reference Materials for Improving Reliability of Multiomics Profiling
Phenomics (2024)
-
Correcting batch effects in large-scale multiomics studies using a reference-material-based ratio method
Genome Biology (2023)
-
Supervised learning of high-confidence phenotypic subpopulations from single-cell data
Nature Machine Intelligence (2023)
-
A comprehensive single-cell map of T cell exhaustion-associated immune environments in human breast cancer
Nature Communications (2023)