Abstract
Stable isotopes are often used to provide an indication of the trophic level (TL) of species. TLs may be derived by using food-web-specific enrichment factors in combination with a representative baseline species. It is challenging to sample stable isotopes for all species, regions and seasons in Arctic ecosystems, e.g. because of practical constraints. Species-specific TLs derived from a single region may be used as a proxy for TLs for the Arctic as a whole. However, its suitability is hampered by incomplete knowledge on the variation in TLs. We quantified variation in TLs of Arctic species by collating data on stable isotopes across the Arctic, including corresponding fractionation factors and baseline species. These were used to generate TL distributions for species in both pelagic and benthic food webs for four Arctic areas, which were then used to determine intra-sample, intra-study, intra-region and inter-region variation in TLs. Considerable variation in TLs of species between areas was observed. This is likely due to differences in parameter choice in estimating TLs (e.g. choice of baseline species) and seasonal, temporal and spatial influences. TLs between regions were higher than the variance observed within regions, studies or samples. This implies that TLs derived within one region may not be suitable as a proxy for the Arctic as a whole. The TL distributions derived in this study may be useful in bioaccumulation and climate change studies, as these provide insight in the variability of trophic levels of Arctic species.
Similar content being viewed by others
Introduction
Food web and food chain models are generally constructed to evaluate the trophic structure and dynamics of ecological communities, typically serving one of three objectives: (1) defining consistent trophic links and patterns within ecological communities, (2) determining factors affecting or structuring these communities and (3) determining flow pathways of contaminants, nutrients and energy through communities and ecosystems (Vander Zanden and Rasmussen 1996). Studies focusing on these objectives predominantly use the trophic level (TL) concept to group species into discrete levels to describe the organism’s position in a food web (Lindeman 1942). However, the use of discrete trophic levels fails to acknowledge the complex interactions between organisms within a food web (Vander Zanden and Rasmussen 1996). Furthermore, use of an average TL per species is controversial as this ignores the variability induced by differences in feeding patterns between individuals within one species (Reed et al. 2016). This is especially the case in Arctic marine systems which exhibit high spatiotemporal intra-species variability in trophic level, e.g. driven by seasonal fluctuations in light and temperature (de Laender et al. 2009). High peaks in abundance of primary producers and declining prey availability due to loss of sea ice in summer lead to changing trophic interactions among species in high-latitude marine environments, affecting the trophic position of species (Murphy et al. 2016; Durant et al. 2019). These fluctuations affect the species’ lipid content and body size, as well as the community composition and related prey availability throughout the year (de Laender et al. 2009; Falk-Petersen et al. 2009; Blanchard et al. 2017).
Traditionally, the trophic level of a species is quantified based on stomach content analysis, which may be hampered by difficulties in identifying organic material and inter- and intra-species differences in digestion rate (Vinagre et al. 2012). Furthermore, stomach content analysis only provides information on what is eaten within a limited time frame and may not be representative of the species’ diet over a longer period (Kiljunen et al. 2006; Vinagre et al. 2012). In contrast, stable isotopic techniques provide a time-weighted measure of trophic level (Post 2002; Carscallen et al. 2012). The isotopic nitrogen signature (δ15N) of a consumer is systematically enriched by 2–4% relative to the consumer’s diet (Post 2002), making δ15N a reliable proxy to describe flows of contaminants and energy through food chains (Walters et al. 2008). Recent studies support the use of a combined approach where the trophic level is inferred from δ15N analysis by using a combination of species-specific and tissue-specific enrichment factors and an ecosystem-specific baseline species. This allows for ecosystem, tissue- and species-specific fractionation, giving a more realistic view of the species’ diet over time than when looking at discrete “actual” trophic levels (de Laender et al. 2009). Although δ15N is considered to accurately describe the trophic level of a species within one region and season, it is considered less accurate when comparing between multiple systems (Vander Zanden et al. 1999) because of the uncertainties introduced by variability in environmental conditions and in feeding habits of species between regions.
Variation in the trophic level of a species—as estimated based on stable nitrogen isotopes—may result from different sources, propagating through different levels of aggregation. These include differences in and between individual organisms, differences in environmental conditions, methodological differences (e.g., assumed baseline species) and analytical errors. Examples include differences in nutrient uptake (notably nitrate) by phytoplankton as a result of differing abiotic circumstances (in, e.g. sea water temperature and sea ice coverage) between Arctic regions may alter the isotopic signature of baseline species used in estimating trophic positions (Tamelander et al. 2009). Moreover, variation may occur in the systematically enrichment of δ15N, depending on the sample tissue, because of tissue turnover rates and metabolic discrimination (Clark et al. 2019). In the present study, we aim to investigate and quantify these sources of variation in the trophic level of Arctic species by aggregating our data across four levels in a ‘variance model’. By determining the variance in TLs on an intra-sample, intra-study, intra-region and inter-region level, we aim to evaluate the utility of using TL estimates for a single region as a proxy for the Arctic as a whole. As the Arctic food-web structure correlates strongly with environmental gradients (Kortsch et al. 2019), we expect variation in TLs at the inter-region level to be higher than variation at the intra-sample, intra-study and intra-region level. To evaluate variability in trophic levels of Arctic species, we collated data on trophic levels, stable nitrogen isotopes, baseline species and fractionation parameters from different studies on multiple Arctic species in both pelagic and benthic food webs.
Materials and methods
Data collection and compilation
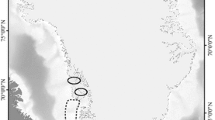
Data on stable nitrogen isotopes for Arctic species, and corresponding TL parameters (see Eq. 1; δ15NBaseline, TLbaseline, Δ15N, ΔD), were collected by conducting an extensive literature search using the Web of Science (Clarivate Analytics 2020). We combined search strings related to stable isotope analysis (e.g. ‘stable isotope analysis’ and ‘nitrogen stable isotopes’) and biota in the European, Canadian and Alaskan Arctic using general terms (e.g. ‘Arctic biota’) as well as species names (e.g. ‘Ursus maritimus’ and ‘Calanus hyperboreus’). Stable isotope data were either extracted from tables or manually digitized using DigitizeIt (http://www.DigitizeIt.de/). Only stable isotope data sampled from April until October and after the year 2000 were included in the dataset, due to a lack of data outside this timeframe. Distinction was made between benthic and pelagic food webs. Although additional organism-specific and sample-specific parameters (i.e. age, length, body weight, date and sampling tissue) were included in the database, no further sub-setting was based on these parameters. The initial search resulted in 65 useful articles and reports, encompassing 148 species, covering four distinct Arctic areas: Alaskan Beaufort Sea, Canadian Beaufort Sea, Canadian Archipelago and Svalbard (Fig. 1; see SI for the full reference list). Data pertaining to unique species (i.e. only observed in one of the four areas) were disregarded, resulting in a dataset comprising 107 species (29 pelagic, 78 benthic species), covering approximately 2400 individual records (see TL dataset.csv in the Supporting Information for raw data (ESM_1.csv)).
Sampling regions of stable nitrogen isotopes in Arctic biota, obtained from an extensive literature search for the Svalbard Archipelago, the Canadian Archipelago, the Canadian Beaufort Sea and the Alaskan Beaufort Sea/Chukchi Sea. The size of each point reflects the relative number of samples at that location
Trophic level estimation and quantifying trophic variation among four Arctic areas
For each record, nitrogen-derived continuous trophic levels were calculated based on nitrogen stable isotope data collected for Arctic biota (δ15N), according to the following general model (Post 2002; Jardine et al. 2006):
where TLconsumer represents the tropic level of the consumer, TLbaseline represents the trophic level of the baseline species for a particular region and study (primary consumer, typically C. hyperboreus), δ15Nconsumer the measured stable nitrogen isotope of the consumer, δ15Nbaseline the measured stable nitrogen isotope of the baseline species, and ∆15N represents the nitrogen enrichment (or fractionation) factor of δ15N across trophic levels. In some cases, the study included an additional diet-tissue isotopic fractionation factor (\(\Delta D\)), correcting for differences in stable nitrogen isotopic values across tissue types (Hobson and Clark 1992). In a minority of the studies, two baseline species were used in calculating the trophic level of a species. In this case, δ15Nbaseline was calculated according to Post (2002):
where δ15Nbaseline, 1 and δ15Nbaseline, 2 represent the measured stable nitrogen isotope of the two baseline species and α represents the reported proportion of the two food sources. If only one baseline species was reported, an α of 1 was assumed.
We used the baseline species, the corresponding δ15Nbaseline values, and the enrichment factors as reported in the associated studies. When comparing between regions, to correct for choice of baseline species, we normalized the average baseline δ15N, standardizing for particulate organic matter (TL = 1) using Eq. 1.
Aggregation of trophic level estimates
In order to compare trophic level estimates between regions, the estimates were aggregated at the level of samples, studies and regions, respectively (Fig. 2). At the sample level, variation due to multiple replicates was quantified by means of Monte Carlo simulation (1000 iterations), i.e. parameters of Eq. 1 for which a mean value and a standard deviation were reported based on multiple replicates (e.g. δ15Nconsumer, δ15Nbaseline and ∆15N) were replaced by normal distributions. The average variance of studies with multiple replicates was used as an indicator of intra-sample variability. At the study and region level, the weighted arithmetic mean and variance of the available average TL values were calculated, using the sample size of the replicates (study level) and samples (region level) as weights. Before aggregation, normality of the average TL values was checked using the Shapiro–Wilk test, and Levene’s test was performed to verify equal variances. For each species and region, the aggregated mean TL and its standard deviation were then used to derive the 2.5th, 50th and 97.5th percentiles of the TL distributions. To account for bias due to differences in species composition between regions, we then compared TL averages based on common species between the four regions by performing an one-way ANOVA.
Species-specific TL estimates were aggregated at different levels, i.e. replicates (R) were aggregated at the sample level, samples (S) were aggregated at the study level, and studies (St) were aggregated at the region level. Finally, species-specific TL estimates at the region level (Rg) were compared
Uncertainty and variability in trophic level estimates
Variation at lower aggregation levels propagates into higher aggregation levels (Fig. 2) according to the following equation (e.g. Wallace and Williams 2005):
where σmeasured,i2 is the variance measured at aggregation level i, VARi is the unbiased variation at level i, \(\overline{{{{\sigma }_{{\rm{measured}}, i-1}}^{2}}}\) is the average measured variance at aggregation level i − 1 and Ni−1 is the sample size at level i − 1. This equation was rearranged to obtain the unbiased variation at each aggregation level:
By comparing the average (unbiased) variation calculated at the intra-sample, intra-study and intra-region level with the (unbiased) variation among the four regions (inter-region), we evaluated the magnitude of different sources of variation and uncertainty. By comparing region TL medians, we could evaluate the utility of using TL estimates from a single region as a proxy for the Arctic as a whole. Additionally, the coefficient of variation (CV) of each TL distribution was calculated to assess the dispersion of species’ trophic level relative to the mean observed TL. All analyses and simulations were performed in R statistics v3.4.1 (see the Supporting Information for the code (ESM_2.txt)).
Identifying main external drivers of variation in trophic level
Principal component analysis (PCA) was used to explore patterns between estimated TL values on the one hand and TL parameters (i.e. δ15Nbaseline, TLbaseline and Δ15N) and extraneous factors (δ13C, sex, age, longitude, latitude, sampling month and year) on the other, using the function prcomp in R statistics v3.4.1. Since relative importance is important in PCA analysis, variables were normalized by min–max scaling. The species nitrogen isotopic values (δ15N) were excluded as a possible influential factor in the principal component analysis, as these were directly used in estimating TL estimates. The most influential predictors of trophic level were then identified by standardizing regression coefficients using the Relaimpo package R statistics 3.4.1.
Results
Nitrogen isotopes and fractionation factors measurements
Our database encompassed 107 species, including species from benthic (N = 78) and pelagic (N = 29) food webs. Nitrogen isotopes varied considerably among taxa and sample regions in both food-web types (Fig. 3 graph a and b). Mean stable nitrogen isotopes (δ15N) per species ranged from 4.8‰ in pelagic particulate organic matter (pelagic-POM) to 20.1‰ in polar bears (U. maritimus) in pelagic food webs and from 3.9‰ (benthic-POM) to 15.2‰ for the eelpout Lycodes polaris in benthic food webs. When looking at inter-regional variation, benthic species sampled at Svalbard on average had significantly lower δ15N values than the same species sampled in Alaska, the Canadian Beaufort Sea and Canadian Archipelago (P = 0.004, F = 7.62, LSD-test). The same holds for pelagic food webs, in which species sampled at Svalbard had significantly lower nitrogen isotopes than their North American counterparts (P < 0.0001, F = 92.13, LSD-test).
Distribution of stable nitrogen isotopes (a and b) and TLs (c and d) and corresponding 95% confidence intervals, for species within the European Arctic (green), the Canadian Beaufort Sea (in blue), the Canadian Archipelago (in red) and Alaskan Arctic (black) for pelagic food webs (left graphs) and benthic food webs (right graphs). Levels of significance when comparing the trophic level of species between regions: ***P < 0.005, **P < 0.01, *P < 0.05, n.s. no significant difference
Table 1 summarizes the stable nitrogen isotope fractionation factors (∆15N) reported in the literature. For pelagic food webs, the fractionation factors differed significantly between regions (P < 0.001, One-way ANOVA/LSD), with highest values reported for Alaska (3.74 ± 0.14, see Table 1) and lowest for the Canadian Archipelago (2.80 ± 0.48). In benthic food webs, ∆15N values did not differ significantly between regions (P = 0.145, One-way ANOVA), with an average fractionation factor of 3.43% (± 0.1).
Baseline species
The majority of TLbaselines reported in the literature were based on Calanus sp. (TL = 2), accounting for 71.5% and 77.7% of all baselines in both benthic and pelagic food webs, respectively (See Table S1 for a complete list of baseline species per study and region). The average trophic level of baseline species reported for pelagic food webs in Alaska was relatively high (3.38 ± 0.98), which can be explained by the use of Phoca hispida as a baseline species by Rogers et al. (2015). Omitting these data records would result in an average TLbaseline for Alaskan pelagic food webs of 1.88 ± 0.5. In benthic food webs, TLbaselines used in Alaska and Svalbard showed to be significantly lower than those used in studies carried out in the Canadian Archipelago and Canadian Beaufort Sea (P < 0.0001, LSD-test). The average baseline δ15N, when standardized for particulate organic matter (TL = 1), among all regions for benthic food webs was 4.94 ± 1.9‰ (Table 1), with the lowest average baseline δ15N reported in Svalbard (4.2 ± 1.14‰) and the highest average baseline δ15N reported in the Canadian Archipelago (5.6 ± 1.64‰). Significant differences in baseline δ15N, standardized for TL = 1, were observed between Svalbard and all other regions in both the pelagic and benthic food web (P < 0.0001, LSD-test, Figure S2). In pelagic food webs, the average standardized baseline δ15N among all regions was 5.61 ± 1.82‰, with the lowest average baseline δ15N again determined for Svalbard (4.94 ± 2.59) and the highest baseline reported for Alaska (5.85 ± 1.18; Table 1 and Figure S2). In benthic food webs, the average standardized baseline δ15N was 4.9 (± 1.90), with the lowest average baseline reported for Svalbard (4.2 ± 1.14) and the highest average baseline reported for the Canadian Archipelago (5.6 ± 1.64).
Trophic level estimates
Figure 3 (lower graphs) shows the calculated species-specific trophic level distributions for pelagic and benthic food webs in the four geographical areas. All TL distributions were normally distributed (P > 0.05, Shapiro–Wilk test) and Levene’s test showed no significant differences in variation between/within regions (P > 0.05). For 21 of the 29 pelagic species in the pelagic food web, statistically significant differences in TLs were observed when comparing TLs between regions (P < 0.05, One-way ANOVA). For benthic food webs, statistically significant differences in TLs were observed for 59 of the 78 species. Table 2 summarizes the TL estimates by reporting the average TL per species per region and per food web. Highest average TLs, based on the average of all species weighted averages, were calculated for Alaska and the Canadian Beaufort Sea in pelagic food webs (3.00 and 2.98, Table 2), and for Alaska and Svalbard in benthic food webs (3.00 and 2.95). The lowest averaged TLs are calculated for the Canadian Archipelago (2.73) and Svalbard (2.50) for pelagic and benthic food webs, respectively. Statistically significant differences were observed in species average TLs between the areas for both the pelagic and benthic food web (P = 0.027, mANOVA, See Tables S2 and S3 for all species averages for δ15N and TLs, respectively).
Variability in trophic level estimates
Figure 4 shows the mean coefficients of variation (CVs) of the TL estimates at the intra-sample, intra-study, intra-region and inter-region level, i.e. averaged over all species. In general, CVs increased with aggregation level, with the smallest CV pertaining to intra-sample variation and the largest CV to inter-region variation. One exception was the pelagic food web of the Svalbard Archipelago, where the highest mean CV was calculated for intra-region variation. Furthermore, the intra-sample CV of benthic food webs in Alaska and the Canadian Archipelago was higher than the intra-study CV. Variances and CVs calculated for all individual species in all four regions are shown in Figs. S4, S5 and S6, S7 in the Supporting Information, respectively (ESM_3.pdf).
Identifying drivers of variation in species’ trophic level
The first two principle components (PC1 and PC2) explained 48.7% of the variation in TLs within the pelagic food-web data and 34.8% in the benthic food-web data (Fig. 5). Components PC3 and PC4 together explained 18.2% and 23% of the variation in the pelagic and benthic food web, respectively. Looking at the correlation between variables included in the principal component analysis revealed that both longitude and sampling month showed to be negatively correlated with trophic position, implying that pelagic organisms sampled at a higher longitude (or sampled later in the year) showed to have a lower trophic position. For latitude, no such correlation with trophic level was found for pelagic food webs. In contrast, in benthic systems, latitude showed to be correlated with trophic level, albeit it a weak correlation. In the PCA plot for the pelagic food web, no correlation between baseline parameters (baseline trophic level and baseline nitrogen isotopic value) and fractionation factor was observed, implying that, given Eq. 1, little variation was observed within used fractionation factors. A strong overlap of the ellipses of the Canadian Arctic and the Svalbard (the European Arctic) in both PCA biplots was observed, indicating a strong uniformity of these regions. In the benthic food web, only longitude was negatively correlated with PC1. However, none of the factor loadings exceeded 0.5, implying that none of the variables have a strong correlation with the principle components. PC2 was negatively correlated with latitude and longitude and the nitrogen isotopic signature in the benthic food web (loadings > 0.4). In the pelagic food web, PC2 was negatively correlated with the trophic level of the baseline species and its nitrogen isotope. However, again none of the factor loadings exceeded 0.5 (Table S4, Supporting Information).
In both food-web types, PC1 and PC2 showed a strong significant discrimination between Alaska and Svalbard. In pelagic food webs, biota sampled from the Alaskan Arctic had lower PC1 values than biota sampled from Svalbard, which was likely mainly attributed to differences in longitude. In addition, biota sampled in Alaskan pelagic food webs had lower nitrogen isotopes than biota from Svalbard. In benthic food webs, the discrimination between Svalbard and Alaska could be mainly explained by the variation in PC2 axis. Again, nitrogen isotopic ratios of the baseline species and its corresponding trophic level in general were lower in biota in Alaskan benthic food webs.
Assessing the relative importance of predictors of trophic level, the pelagic food web showed female sex (6.4%) and baseline δ15N (5.75%) to be the strongest predictors, after the reported stable nitrogen isotope itself. The relative importance of TL parameters, notably parameters associated with baseline species, generally was higher than the relative importance of extraneous variables (e.g. sampling year or region; Fig. S3 and Table S5, Supporting Information). One notable exception is sampling month (5.3%), suggesting strong seasonality of trophic levels of species. In benthic food webs, again baseline species δ15N and baseline trophic level were more important than most of the external variables, with 6.8% and 3.1%, respectively. Sampling year showed to be the strongest external predictor of trophic level in benthic food webs, which may be due to inter-annual fluctuations in environmental conditions influencing plankton composition in Arctic oceans (Batten et al. 2018). The pelagic food web confirmed the variables month (6.5%) and sex (6.1%) as the strongest predictors of trophic level, after the stable nitrogen isotopic values itself, showing these variables to be a stronger predictor for trophic level than any of the trophic level parameters.
Discussion
Trophic level estimates
On average, the highest average nitrogen isotopes and TLs were calculated for pelagic species sampled in Alaska. When comparing averages of common species, taking into account sampling preference of a certain species, statistically significant differences (P < 0.05, one-way ANOVA) were observed between regions for both food-web types. Median trophic levels, based on species averages reported in this study, are similar to the median nitrogen isotopic baseline-corrected TL (3.17 ± 0.88) and median TL estimates (3.32 ± 0.79) for Arctic areas reported by Carscallen et al. (2012). Furthermore, these values were similar to TL estimates for Arctic areas based on stomach content or fatty acid studies for Arctic marine mammals (Pauly et al. 1998; Trites 2001).
The calculation of trophic levels and its variability to compare TLs of species between regions depend on a number of assumptions. First, we assumed all input parameters (e.g. δ15Nconsumer, δ15Nbaseline and ∆15N) not to be correlated, as a positive correlation between these parameters may result in an underestimation of the variation in estimated TLs. However, a correlation test revealed that none of these parameters were correlated (r < 0.1). Our second assumption pertains to the use of an accurate nitrogen fractionation factor (Δ15N). In this study, all fractionation factors applied in estimating TLs of Arctic species in benthic food webs were between 3.4 and 3.8. In contrast, Δ15Ns used in studies focusing on pelagic food webs were much lower, which is due to the low factor applied in ringed seals (P. hispida) in multiple studies in the Canadian Archipelago (2.4‰) (Brown et al. 2016; Houde et al. 2017). Although the majority of studies included in the present study use fractionation constants as reported by Post (2002) (3.4) or Hobson and Welch (1992) (3.8), a wide range of fractionation factor values were reported in the literature (Lecomte et al. 2011; L’Hérault et al. 2018). Multiple studies revealed that the nitrogen fractionation factor Δ15N may decrease with increasing dietary protein quality in the organisms diet, implying that nitrogen discrimination will be lower for species at higher trophic levels (i.e. carnivores) than for herbivorous or omnivorous species (Robbins et al. 2005, 2010; Florin et al. 2011). Next to protein quality, protein quantity may also play an important role in variation in nitrogen fractionation, although this relationship showed to be very complex (Florin et al. 2011; Hughes et al. 2018). As enrichment of δ15N may be highly influenced by temperature, growth rate, dietary protein quality and sampling tissue (Lecomte et al. 2011; Clark et al. 2019), estimates derived from experiments may not reflect factors as observed in natural settings. However, the lack of understanding of sources of variation in Δ15N within and between species and its lack of experimental validation is very common in the field of wildlife studies (Morrissey et al. 2004; Lecomte et al. 2011). To account for intra-population variation in Δ15N, there is a strong need for unbiased, statistically valid taxa and tissue-specific estimates (Vanderklift and Ponsard 2003; Auerswald et al. 2010; Hussey et al. 2014).
A third assumption in this study is that species δ15N have been sampled and measured correctly. Tissue samples exposed to air tend to degrade, possibly enriching δ15N between 0.6 and 1.3‰ (Perkins et al. 2018). Furthermore, chemical lipid extraction techniques to obtain lipid-free δ15N may sometimes affect measured stable nitrogen isotopes, as amino acids may leach from the tissue, reducing the accuracy of the TL estimates (Sotiropoulos et al. 2004; Clark et al. 2019). However, δ15N is assumed to increase with 0.3–0.5‰ due to lipid removal, which is relatively small compared to the typical fractionation factor used in many studies in trophic ecology (Sotiropoulos et al. 2004). In the present study, variance induced by lipid removal techniques across studies pertained to intra-regional variance.
Our final assumption is that the temporal and spatial scale at which we aggregated that the stable isotope data are sufficiently small enough to exclude major variation induced by differences between ecosystems. Individual TL estimates were based on associated baseline species and δ15Nbaseline, including variation in basal resources, sampled within the same study and site. Nevertheless, inappropriately aggregating data at too large a scale may lead to introducing bias in the derived TL probability distributions, due to for instance different offsets of algae blooming at higher latitudes, potentially leading to different results and conclusions (Falk-Petersen et al. 2009; Nielsen et al. 2018).
Variability in trophic level estimates
In general, the variance calculated between the regions was higher than the variance between samples and studies within one region, for both pelagic and benthic food webs. This is especially the case for benthic species, for which the overall mean calculated variance between regions was a factor 2.5 higher than the variance determined for individuals, samples and studies.
Identifying drivers of variation in species’ trophic level
In both food webs, in general, TL parameters (notably, parameters related to baseline species: TLbaseline and δ15Nbaseline) were stronger correlated with TL estimates than any of the other included variables, with one notable exception being sampling month. These findings are in line with the generally adopted idea of the choice of an appropriate baseline being the most important in trophic studies using stable nitrogen isotopes, regardless of the research question (Post 2002). TLs showed to be highly correlated with the stable nitrogen isotopes of the baseline species in both food webs. This again underlines the importance of choice of baseline species. Unlike in the benthic food web, the TL of species in the pelagic food web was strongly correlated to sampling month. This suggests that seasonality in trophic levels may be larger in the species from pelagic food webs than for benthic species. This may particularly be the case for the polar bear, covering a substantial part of our dataset, a species well known for its seasonality in feeding habits (Iversen et al. 2013). As sea ice declines in summer, a fraction of polar bears following the retreat of ice beyond the continental shelve may get food deprived, as they switch to alternative food sources residing at lower trophic levels, resulting in lower measured δ15N values (Whiteman et al. 2018). Furthermore, in pelagic marine systems, seasonal changes in lipid content of pelagic baseline species occur due to negligible light in winter and changing POM composition and algae blooms in spring and summer. This may result in differences in isotopic signatures of zooplankton across seasons, especially when comparing between studies where researchers did not remove lipids prior to isotopic analysis (Hobson and Welch 1992; Søreide et al. 2006). Sex showed to be a relatively strong predictor of TL in pelagic food webs in the present study (6.1%), with females showing higher average TLs than males. This may be due to the relative large proportion of mammalian marine predators in our dataset. Multiple studies (Hobson et al. 1997; Lowry et al. 1980; Dehn et al. 2007; Thiemann et al. 2008) support our findings and report higher δ15N in female marine mammals and/or find significant differences in prey composition between sexes (with females being more carnivorous than males). Dehn et al. (2007) propose that these differences in prey composition in seals may be partially explained by spatial segregation and differential use of resources, which is also visible in polar bears during the breeding season (Laidre et al. 2013).
Although it is known that the life cycle of pelagic zooplankton species shorten with latitude (Lischka and Hagen 2007), no strong correlation was found between latitude and TLs in the present study. An additional potential predictor of trophic level includes body size. Although this predictor was not included in the present study, due to lack of data, a strong correlation between body size and trophic level was acknowledged in multiple recent studies, especially for fish and mammals (Lesage et al. 2001; Landry et al. 2018).
Recommendations and implications
To our knowledge, the present study was the first to quantify variation of trophic levels of species across the marine Arctic. Unfortunately, many studies in trophic ecology rely on small numbers of stable isotope data, scattered throughout the time and space, due to financial and logistical constraints (Ward et al. 2010), making it challenging to quantify variation in TLs at one time and region. Nevertheless, multiple studies did account for variation by means of experimental design (e.g. Bayesian inference), similar to methods proposed in the present study (Starrfelt et al. 2013; Kim et al. 2016). Generating TL distributions based on occurring variation in δ15N among species, as well as methodological variation in TL parameters, rather than using a single discrete TL value for one species, allows for a better understanding of feeding behaviour of species in the Arctic. Understanding trophodynamics across all trophic levels and scales in Arctic areas is highly important in modelling impacts of anthropogenic drivers (Loseto et al. 2009; Cherry et al. 2011). TL distributions may be useful in providing naturally realistic predictions in bioaccumulation or climate change studies, as these are based on the probability of a species situating at a certain trophic level. Therefore, a logical next step is to apply these distributions in bioaccumulation models, to evaluate their implications in actual modelling practice.
References
Auerswald K, Wittmer MH, Zazzo A, Schäufele R, Schnyder H (2010) Biases in the analysis of stable isotope discrimination in food webs. J Appl Ecol 47:936–941. https://doi.org/10.1111/j.1365-2664.2009.01764.x
Batten SD, Raitsos DE, Danielson S, Hopcroft R, Coyle K, McQuatters-Gollop A (2018) Interannual variability in lower trophic levels on the Alaskan Shelf. Deep Sea Res Part II 147:58–68. https://doi.org/10.1016/j.dsr2.2017.04.023
Blanchard AL, Day RH, Gall AE, Aerts LA, Delarue J, Dobbins EL, Wisdom SS (2017) Ecosystem variability in the offshore northeastern Chukchi Sea. Prog Oceanogr 159:130–153. https://doi.org/10.1016/j.pocean.2017.08.008
Brown TM, Fisk AT, Wang X, Ferguson SH, Young BG, Reimer KJ, Muir DC (2016) Mercury and cadmium in ringed seals in the Canadian Arctic: influence of location and diet. Sci Total Environ 545:503–511. https://doi.org/10.1016/j.scitotenv.2015.12.030
Carscallen WMA, Vandenberg K, Lawson JM, Martinez ND, Romanuk TN (2012) Estimating trophic position in marine and estuarine food webs. Ecosphere 3:1–20. https://doi.org/10.1890/ES11-00224.1
Cherry SG, Derocher AE, Hobson KA, Stirling I, Thiemann GW (2011) Quantifying dietary pathways of proteins and lipids to tissues of a marine predator. J Appl Ecol 48:373–381. https://doi.org/10.1111/j.1365-2664.2010.01908.x
Clarivate Analytics (2020) Web of Science Core Collection. Retrieved from www.webofknowledge.com
Clark CT, Horstmann L, Misarti N (2019) Lipid normalization and stable isotope discrimination in Pacific walrus tissues. Sci Rep 9(1):5843
de Laender F, van Oevelen D, Frantzen S, Middelburg JJ, Soetaert K (2009) Seasonal PCB bioaccumulation in an Arctic marine ecosystem: a model analysis incorporating lipid dynamics, food-web productivity and migration. Environ Sci Technol 44:356–361. https://doi.org/10.1021/es902625u
Dehn L, Sheffield G, Follmann E, Duffy L, Thomas DL, O’Hara T (2007) Feeding ecology of phocid seals and some walrus in the Alaskan and Canadian Arctic as determined by stomach contents and stable isotope analysis. Polar Biol 30(2):167–181
Durant JM, Molinero J-C, Ottersen G, Reygondeau G, Stige LC, Langangen Ø (2019) Contrasting effects of rising temperatures on trophic interactions in marine ecosystems. Sci Rep 9(1):1–9
Falk-Petersen S, Mayzaud P, Kattner G, Sargent JR (2009) Lipids and life strategy of Arctic Calanus. Mar Biol Res 5:18–39. https://doi.org/10.1080/17451000802512267
Florin ST, Felicetti LA, Robbins CT (2011) The biological basis for understanding and predicting dietary-induced variation in nitrogen and sulphur isotope ratio discrimination. Funct Ecol 25:519–526. https://doi.org/10.1111/j.1365-2435.2010.01799.x
Hobson KA, Clark RG (1992) Assessing avian diets using stable isotopes II: factors influencing diet-tissue fractionation. Condor. https://doi.org/10.2307/1368808
Hobson KA, Welch HE (1992) Determination of trophic relationships within a high Arctic marine food web using δ 13 C and δ 15 N analysis. Mar Ecol Prog Ser. https://doi.org/10.3354/meps084009
Hobson KA, Sease JL, Merrick RL, Piatt JF (1997) Investigating trophic relationships of pinnipeds in Alaska and Washington using stable isotope ratios of nitrogen and carbon. Mar Mamm Sci 13(1):114–132
Houde M, Wang X, Ferguson S, Gagnon P, Brown T, Tanabe S, Muir D (2017) Spatial and temporal trends of alternative flame retardants and polybrominated diphenyl ethers in ringed seals (Phoca hispida) across the Canadian Arctic. Environ Pollut 223:266–276. https://doi.org/10.1016/j.envpol.2017.01.023
Hughes KL, Whiteman JP, Newsome SD (2018) The relationship between dietary protein content, body condition, and Δ 15 N in a mammalian omnivore. Oecologia 186:357–367. https://doi.org/10.1007/s00442-017-4010-5
Hussey NE, MacNeil MA, McMeans BC, Olin JA, Dudley SF, Cliff G, Fisk AT (2014) Rescaling the trophic structure of marine food webs. Ecol Lett 17(2):239–250
Iversen M, Aars J, Haug T, Alsos IG, Lydersen C, Bachmann L, Kovacs KM (2013) The diet of polar bears (Ursus maritimus) from Svalbard, Norway, inferred from scat analysis. Polar Biol 36:561–571. https://doi.org/10.1007/s00300-012-1284-2
Jardine TD, Kidd KA, Fisk AT (2006) Applications, considerations, and sources of uncertainty when using stable isotope analysis in ecotoxicology. Environ Sci Technol 40:7501–7511. https://doi.org/10.1021/es061263h
Kiljunen M, Grey J, Sinisalo T, Harrod C, Immonen H, Jones RI (2006) A revised model for lipid-normalizing δ13C values from aquatic organisms, with implications for isotope mixing models. J Appl Ecol 43:1213–1222. https://doi.org/10.1111/j.1365-2664.2006.01224.x
Kim J, Gobas FAPC, Arnot JA, Powell DE, Seston RM, Woodburn KB (2016) Evaluating the roles of biotransformation, spatial concentration differences, organism home range, and field sampling design on trophic magnification factors. Sci Total Environ 551–552:438–451. https://doi.org/10.1016/j.scitotenv.2016.02.013
Kortsch S, Primicerio R, Aschan M, Lind S, Dolgov AV, Planque B (2019) Food-web structure varies along environmental gradients in a high-latitude marine ecosystem. Ecography 42(2):295–308. https://doi.org/10.1111/ecog.03443
L’Hérault V, Lecomte N, Truchon MH, Berteaux D (2018) Discrimination factors of carbon and nitrogen stable isotopes from diet to hair in captive large Arctic carnivores of conservation concern. Rapid Commun Mass Spectrom. https://doi.org/10.1002/rcm.8239
Laidre KL, Born EW, Gurarie E, Wiig Ø, Dietz R, Stern H (2013) Females roam while males patrol: divergence in breeding season movements of pack-ice polar bears (Ursus maritimus). Proc R Soc B Biol Sci 280(1752):20122371
Landry JJ, Fisk AT, Yurkowski DJ, Hussey NE, Dick T, Crawford RE, Kessel ST (2018) Feeding ecology of a common benthic fish, shorthorn sculpin (Myoxocephalus scorpius) in the high arctic. Polar Biol. https://doi.org/10.1007/s00300-018-2348-8
Lecomte N, Ahlstrøm Ø, Ehrich D, Fuglei E, Ims RA, Yoccoz NG (2011) Intrapopulation variability shaping isotope discrimination and turnover: experimental evidence in arctic foxes. PLoS One 6:e21357. https://doi.org/10.1371/journal.pone.0021357
Lesage V, Hammill MO, Kovacs KM (2001) Marine mammals and the community structure of the Estuary and Gulf of St Lawrence, Canada: evidence from stable isotope analysis. Mar Ecol Prog Ser 210:203–221. https://doi.org/10.3354/meps210203
Lindeman RL (1942) The trophic-dynamic aspect of ecology. Ecology 23:399–417. https://doi.org/10.2307/1930126
Lischka S, Hagen W (2007) Seasonal lipid dynamics of the copepods Pseudocalanus minutus (Calanoida) and Oithona similis (Cyclopoida) in the Arctic Kongsfjorden (Svalbard). Mar Biol 150:443–454. https://doi.org/10.1007/s00227-006-0359-4
Loseto L, Stern G, Connelly T, Deibel D, Gemmill B, Prokopowicz A, Ferguson S (2009) Summer diet of beluga whales inferred by fatty acid analysis of the eastern Beaufort Sea food web. J Exp Mar Biol Ecol 374:12–18. https://doi.org/10.1016/j.jembe.2009.03.015
Lowry LF, Frost KJ, Burns JJ (1980) Variability in the diet of ringed seals, Phoca hispida, in Alaska. Can J Fish Aquat Sci 37(12):2254–2261
Morrissey CA, Bendell-Young LI, Elliott JE (2004) Linking contaminant profiles to the diet and breeding location of American dippers using stable isotopes. J Appl Ecol 41:502–512. https://doi.org/10.1111/j.0021-8901.2004.00907.x
Murphy EJ, Cavanagh RD, Drinkwater KF, Grant SM, Heymans J, Hofmann EE, Johnston NM (2016) Understanding the structure and functioning of polar pelagic ecosystems to predict the impacts of change. Proc R Soc B Biol Sci 283(1844):20161646
Nielsen JM, Clare EL, Hayden B, Brett MT, Kratina P (2018) Diet tracing in ecology: method comparison and selection. Methods Ecol Evol 9:278–291. https://doi.org/10.1111/2041-210X.12869
Pauly D, Trites A, Capuli E, Christensen V (1998) Diet composition and trophic levels of marine mammals. ICES J Mar Sci 55:467–481. https://doi.org/10.1006/jmsc.1997.0280
Perkins MJ, Mak YK, Tao LS, Wong AT, Yau JK, Baker DM, Leung KM (2018) Short-term tissue decomposition alters stable isotope values and C: N ratio, but does not change relationships between lipid content, C: N ratio, and Δδ13C in marine animals. PLoS One 13:e0199680. https://doi.org/10.1371/journal.pone.0199680
Post DM (2002) Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology 83:703–718. https://doi.org/10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2
Reed J, Shannon L, Velez L, Akoglu E, Bundy A, Coll M, Shin Y-J (2016) Ecosystem indicators—accounting for variability in species’ trophic levels. ICES J Mar Sci 74:158–169
Robbins CT, Felicetti LA, Sponheimer M (2005) The effect of dietary protein quality on nitrogen isotope discrimination in mammals and birds. Oecologia 144(4):534–540. https://doi.org/10.1007/s00442-005-0021-8
Robbins CT, Felicetti LA, Florin ST (2010) The impact of protein quality on stable nitrogen isotope ratio discrimination and assimilated diet estimation. Oecologia 162:571–579
Rogers M, Peacock E, Simac K, O’Dell M, Welker J (2015) Diet of female polar bears in the southern Beaufort Sea of Alaska: evidence for an emerging alternative foraging strategy in response to environmental change. Polar Biol 38:1035–1047. https://doi.org/10.1007/s00300-015-1665-4
Søreide JE, Hop H, Carroll ML, Falk-Petersen S, Hegseth EN (2006) Seasonal food web structures and sympagic–pelagic coupling in the European Arctic revealed by stable isotopes and a two-source food web model. Prog Oceanogr 71:59–87. https://doi.org/10.1016/j.pocean.2006.06.001
Sotiropoulos MA, Tonn WM, Wassenaar LI (2004) Effects of lipid extraction on stable carbon and nitrogen isotope analyses of fish tissues: potential consequences for food web studies. Ecol Freshw Fish 13(3):155–160
Starrfelt J, Borgå K, Ruus A, Fjeld E (2013) Estimating trophic levels and trophic magnification factors using Bayesian inference. Environ Sci Technol 47:11599–11606. https://doi.org/10.1021/es401231e
Tamelander T, Kivimäe C, Bellerby RGJ, Renaud PE, Kristiansen S (2009) Base-line variations in stable isotope values in an Arctic marine ecosystem: effects of carbon and nitrogen uptake by phytoplankton. Hydrobiologia 630:63–73. https://doi.org/10.1007/s10750-009-9780-2
Thiemann GW, Iverson SJ, Stirling I (2008) Polar bear diets and arctic marine food webs: insights from fatty acid analysis. Ecol Monogr 78(4):591–613
Trites A (2001) Marine mammal trophic levels and interactions. In: Steele JH (ed) Encyclopedia of ocean sciences, 2nd edn. Academic Press, pp 622–627
Vander Zanden MJ, Rasmussen JB (1996) A trophic position model of pelagic food webs: impact on contaminant bioaccumulation in lake trout. Ecol Monogr 66:451–477. https://doi.org/10.2307/2963490
Vander Zanden MJ, Shuter BJ, Lester N, Rasmussen JB (1999) Patterns of food chain length in lakes: a stable isotope study. Am Nat 154:406–416. https://doi.org/10.1086/303250
Vanderklift MA, Ponsard S (2003) Sources of variation in consumer-diet δ 15 N enrichment: a meta-analysis. Oecologia 136:169–182. https://doi.org/10.1007/s00442-003-1270-z
Vinagre C, Salgado JP, Mendonça V, Cabral H, Costa MJ (2012) Isotopes reveal fluctuation in trophic levels of estuarine organisms, in space and time. J Sea Res 72:49–54. https://doi.org/10.1016/j.seares.2012.05.010
Wallace L, Williams R (2005) Validation of a method for estimating long-term exposures based on short-term measurements. Risk Anal Int J 25(3):687–694
Walters DM, Fritz KM, Johnson BR, Lazorchak JM, McCormick FH (2008) Influence of trophic position and spatial location on polychlorinated biphenyl (PCB) bioaccumulation in a stream food web. Environ Sci Technol 42:2316–2322. https://doi.org/10.1021/es0715849
Ward EJ, Semmens BX, Schindler DE (2010) Including source uncertainty and prior information in the analysis of stable isotope mixing models. Environ Sci Technol 44:4645–4650. https://doi.org/10.1021/es100053v
Whiteman JP, Harlow HJ, Durner GM, Regehr EV, Amstrup SC, Ben-David M (2018) Phenotypic plasticity and climate change: can polar bears respond to longer Arctic summers with an adaptive fast? Oecologia 186(2):369–381
Acknowledgements
This research project was funded by the Netherlands Organisation for Scientific Research (NWO), under the Netherlands Polar Programme (NPP), Project No. : 866.13.007. We thank the three anonymous reviewers for their careful reading of our manuscript and their many insightful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hoondert, R.P.J., van den Brink, N.W., van den Heuvel-Greve, M.J. et al. Variability in nitrogen-derived trophic levels of Arctic marine biota. Polar Biol 44, 119–131 (2021). https://doi.org/10.1007/s00300-020-02782-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-020-02782-4