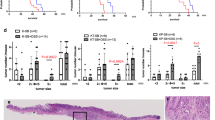
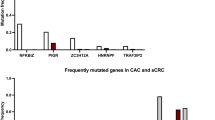
Abstract
Despite advances in the detection and therapy of colorectal cancer (CRC) in recent years, CRC has remained a major challenge in clinical practice. Although alternative methods for modeling CRC have been developed, animal models of CRC remain helpful when analyzing molecular aspects of pathogenesis and are often used to perform preclinical in vivo studies of potential therapeutics. This protocol updates our protocol published in 2007, which provided an azoxymethane (AOM)-based setup for investigations into sporadic (Step 5A) and, when combined with dextran sodium sulfate (Step 5B), inflammation-associated tumor growth. This update also extends the applications beyond those of the original protocol by including an option in which AOM is serially applied to mice with p53 deficiency in the intestinal epithelium (Step 5C). In this model, the combination of p53 deficiency and AOM promotes tumor development, including growth of invasive cancers and lymph node metastasis. It also provides details on analysis of colorectal tumor growth and metastasis, including analysis of partial epithelial-to-mesenchymal transition, cell isolation and co-culture studies, high-resolution mini-endoscopy, light-sheet fluorescence microscopy and micro-CT imaging in mice. The target audience for our protocol is researchers who plan in vivo studies to address mechanisms influencing sporadic or inflammation-driven tumor development, including the analysis of local invasiveness and lymph node metastasis. It is suitable for preclinical in vivo testing of novel drugs and other interventional strategies for clinical translation, plus the evaluation of emerging imaging devices/modalities. It can be completed within 24 weeks (using Step 5A/C) or 10 weeks (using Step 5B).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
All data generated during this study are included in this published article (Fig. 2, Fig. 4a, Fig. 7, Fig. 9b, images in Box 2). Some experiments were performed during previous studies, and similar data were published earlier in different formats (Fig. 3 in ref. 24, Fig. 4b,c and Fig. 5 in ref. 23, Fig. 6 in ref. 20, Fig. 8 and Fig. 9a in ref. 23). Additional information can be provided by the authors upon request.
References
Brenner, H., Kloor, M. & Pox, C. P. Colorectal cancer. Lancet 383, 1490–1502 (2014).
Powell, S. M. et al. Molecular diagnosis of familial adenomatous polyposis. N. Engl. J. Med. 329, 1982–1987 (1993).
Peltomaki, P. Update on Lynch syndrome genomics. Fam. Cancer 15, 385–393 (2016).
Beaugerie, L. & Itzkowitz, S. H. Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 372, 1441–1452 (2015).
Wei, E. K. et al. Comparison of risk factors for colon and rectal cancer. Int. J. Cancer 108, 433–442 (2004).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Koliaraki, V., Pallangyo, C. K., Greten, F. R. & Kollias, G. Mesenchymal cells in colon cancer. Gastroenterology 152, 964–979 (2017).
Hanahan, D. & Coussens, L. M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012).
Zhu, W. et al. Editing of the gut microbiota reduces carcinogenesis in mouse models of colitis-associated colorectal cancer. J. Exp. Med. 216, 2378–2393 (2019).
Neufert, C., Becker, C. & Neurath, M. F. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2, 1998–2004 (2007).
Corpet, D. E. & Pierre, F. How good are rodent models of carcinogenesis in predicting efficacy in humans? A systematic review and meta-analysis of colon chemoprevention in rats, mice and men. Eur. J. Cancer 41, 1911–1922 (2005).
Boivin, G. P. et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology 124, 762–777 (2003).
Moser, A., Pitot, H. & Dove, W. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247, 322–324 (1990).
Roper, J. et al. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat. Protoc. 13, 217–234 (2018).
Fumagalli, A. et al. A surgical orthotopic organoid transplantation approach in mice to visualize and study colorectal cancer progression. Nat. Protoc. 13, 235–247 (2018).
Roper, J. et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017).
Bissahoyo, A. et al. Azoxymethane is a genetic background-dependent colorectal tumor initiator and promoter in mice: effects of dose, route, and diet. Toxicol. Sci. 88, 340–345 (2005).
Stolfi, C. et al. Inhibition of colon carcinogenesis by 2-methoxy-5-amino-N-hydroxybenzamide, a novel derivative of mesalamine. Gastroenterology 138, 221–230 (2010).
Schwitalla, S. et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell 23, 93–106 (2013).
Nambiar, P. R. et al. Preliminary analysis of azoxymethane induced colon tumors in inbred mice commonly used as transgenic/knockout progenitors. Int. J. Oncol. 22, 145–150 (2003).
Suzuki, R., Miyamoto, S., Yasui, Y., Sugie, S. & Tanaka, T. Global gene expression analysis of the mouse colonic mucosa treated with azoxymethane and dextran sodium sulfate. BMC Cancer 7, 84 (2007).
Heichler, C. et al. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut 69, 1269–1282 (2020).
Neufert, C. et al. Tumor fibroblast-derived epiregulin promotes growth of colitis-associated neoplasms through ERK. J. Clin. Invest. 123, 1428–1443 (2013).
Sohn, O. S., Fiala, E. S., Requeijo, S. P., Weisburger, J. H. & Gonzalez, F. J. Differential effects of CYP2E1 status on the metabolic activation of the colon carcinogens azoxymethane and methylazoxymethanol. Cancer Res. 61, 8435–8440 (2001).
Fiala, E. S. Investigations into the metabolism and mode of action of the colon carcinogens 1,2-dimethylhydrazine and azoxymethane. Cancer 40, 2436–2445 (1977).
Reddy, B. S., Weisburger, J. H., Narisawa, T. & Wynder, E. L. Colon carcinogenesis in germ-free rats with 1,2-dimethylhydrazine and N-methyl-Nʹ-nitro-N-nitrosoguanidine. Cancer Res. 34, 2368–2372 (1974).
Evans, J. T., Hauschka, T. S. & Mittelman, A. Brief communication: differential susceptibility of four mouse strains to induction of multiple large-bowel neoplasms by 1,2-dimethylhydrazine. J. Natl Cancer Inst. 52, 999–1000 (1974).
Suzuki, R., Kohno, H., Sugie, S., Nakagama, H. & Tanaka, T. Strain differences in the susceptibility to azoxymethane and dextran sodium sulfate-induced colon carcinogenesis in mice. Carcinogenesis 27, 162–169 (2005).
Tanaka, T. et al. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 94, 965–973 (2003).
De Oliveira, T. et al. Loss of Stat6 affects chromatin condensation in intestinal epithelial cells causing diverse outcome in murine models of inflammation-associated and sporadic colon carcinogenesis. Oncogene 38, 1787–1801 (2019).
Llewellyn, S. R. et al. Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology 154, 1037–1046 e1032 (2018).
Mähler, M. et al. Differential susceptibility of inbred mouse strains to dextran sulfate sodium-induced colitis. Am. J. Physiol. 274, G544–G551 (1998).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Madison, B. B. et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 277, 33275–33283 (2002).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Rausch, P. et al. Analysis of factors contributing to variation in the C57BL/6J fecal microbiota across German animal facilities. Int. J. Med. Microbiol. 306, 343–355 (2016).
Miller, L. R. et al. Considering sex as a biological variable in preclinical research. FASEB J. 31, 29–34 (2017).
Lee, S. M. et al. The effect of sex on the azoxymethane/dextran sulfate sodium-treated mice model of colon cancer. J. Cancer Prev. 21, 271–278 (2016).
Amos-Landgraf, J. M. et al. Sex disparity in colonic adenomagenesis involves promotion by male hormones, not protection by female hormones. Proc. Natl Acad. Sci. USA 111, 16514–16519 (2014).
Crnčec, I. et al. STAT1 is a sex-specific tumor suppressor in colitis-associated colorectal cancer. Mol. Oncol. 12, 514–528 (2018).
Takaku, K. et al. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell 92, 645–656 (1998).
Kucherlapati, M. H. et al. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology 138, 993–1002 e1001 (2010).
O’Rourke, K. P. et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol. 35, 577–582 (2017).
Shibata, H. et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 278, 120–123 (1997).
Drost, J. et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47 (2015).
Zigmond, E. et al. Utilization of murine colonoscopy for orthotopic implantation of colorectal cancer. PloS ONE 6, e28858 (2011).
Vlachogiannis, G. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018).
Mahe, M. M. et al. Establishment of gastrointestinal epithelial organoids. Curr. Protoc. Mouse Biol. 3, 217–240 (2013).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Becker, C., Fantini, M. C. & Neurath, M. F. High resolution colonoscopy in live mice. Nat. Protoc. 1, 2900–2904 (2006).
Neurath, M. F. et al. Assessment of tumor development and wound healing using endoscopic techniques in mice. Gastroenterology 139, 1837–1843 e1831 (2010).
Bialkowska, A. B., Ghaleb, A. M., Nandan, M. O. & Yang, V. W. Improved Swiss-rolling technique for intestinal tissue preparation for immunohistochemical and immunofluorescent analyses. J. Vis. Exp. https://doi.org/10.3791/54161 (2016).
Jung, D. et al. Contrast-enhanced microCT for visualizing and evaluating murine intestinal inflammation. Theranostics 8, 6357–6366 (2018).
Zundler, S. et al. Three-dimensional cross-sectional light-sheet microscopy imaging of the inflamed mouse gut. Gastroenterology 153, 898–900 (2017).
Nieto, M. A., Huang, R. Y., Jackson, R. A. & Thiery, J. P. EMT: 2016. Cell 166, 21–45 (2016).
Stemmler, M. P., Eccles, R. L., Brabletz, S. & Brabletz, T. Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 21, 102–112 (2019).
Isella, C. et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 47, 312–319 (2015).
Baek, S. J. et al. Nonsteroidal anti-inflammatory drug-activated gene-1 over expression in transgenic mice suppresses intestinal neoplasia. Gastroenterology 131, 1553–1560 (2006).
Pallangyo, C. K., Ziegler, P. K. & Greten, F. R. IKKβ acts as a tumor suppressor in cancer-associated fibroblasts during intestinal tumorigenesis. J. Exp. Med. 212, 2253–2266 (2015).
Gage, G. J., Kipke, D. R. & Shain, W. Whole animal perfusion fixation for rodents. J. Vis. Exp. https://doi.org/10.3791/3564 (2012).
Klingberg, A. et al. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J. Am. Soc. Nephrol. 28, 452–459 (2017).
Schmid, B. et al. 3Dscript: animating 3D/4D microscopy data using a natural-language-based syntax. Nat. Methods 16, 278–280 (2019).
Acknowledgements
We thank K. Enderle, K. Hofmann, and I. Zoeller-Utz for excellent technical assistance. This work was funded by the DFG (FOR2438/project 9 to C.N. and M.F.N.; NE1927/2-2, SFB1181-C02 and TRR241-A08 to C.N.) and by the Bartling Stiftung (to C.N.). T.B. was supported by grants from German Research Foundation (FOR 2438/project 4; BR 1399/9-1; 1399/10-1; 1399/13-1).
Author information
Authors and Affiliations
Contributions
C.N., C.H., T.B., K.S. and V.B. designed and performed the experiments. C.N., C.H., T.B., V.B., F.R.G. and M.F.N. analyzed and discussed the data. C.N., C.H. and T.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Jarom Heijmans and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Related links
Key references using this protocol
Neufert, C. et al. J. Clin. Invest. 123, 1428–1443 (2013): https://doi.org/10.1172/JCI63748
Schwitalla, S. et al. Cancer Cell 23, 93–103 (2013): https://doi.org/10.1016/j.ccr.2012.11.014
Heichler, C. et al. Gut 69, 1269–1282 (2020): https://doi.org/10.1136/gutjnl-2019-319200
This protocol is an update to: Nat. Protoc. 2, 1998–2004 (2007): https://doi.org/10.1038/nprot.2007.279
Supplementary information
Rights and permissions
About this article
Cite this article
Neufert, C., Heichler, C., Brabletz, T. et al. Inducible mouse models of colon cancer for the analysis of sporadic and inflammation-driven tumor progression and lymph node metastasis. Nat Protoc 16, 61–85 (2021). https://doi.org/10.1038/s41596-020-00412-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00412-1
This article is cited by
-
Impact of preoperative white blood cell count on outcomes in different stage colorectal cancer patients undergoing surgical resection: a single-institution retrospective cohort study
BMC Cancer (2024)
-
Bifidobacterium adolescentis induces Decorin+ macrophages via TLR2 to suppress colorectal carcinogenesis
Journal of Experimental & Clinical Cancer Research (2023)
-
Predicting response to immunotherapy in gastric cancer via assessing perineural invasion-mediated inflammation in tumor microenvironment
Journal of Experimental & Clinical Cancer Research (2023)
-
Reprogramming of palmitic acid induced by dephosphorylation of ACOX1 promotes β-catenin palmitoylation to drive colorectal cancer progression
Cell Discovery (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.