Abstract
Presence of glycogen granules in anaerobic ammonium-oxidizing (anammox) bacteria has been reported so far. However, very little is known about their glycogen metabolism and the exact roles. Here, we studied the glycogen metabolism in “Ca. Brocadia sinica” growing in continuous retentostat cultures with bicarbonate as a carbon source. The effect of the culture growth phase was investigated. During the growing phase, intracellular glycogen content increased up to 32.6 mg-glucose (g-biomass dry wt)−1 while the specific growth rate and ATP/ADP ratio decreased. The accumulated glycogen begun to decrease at the onset of entering the near-zero growth phase and was consumed rapidly when substrates were depleted. This clearly indicates that glycogen was synthesized and utilized as an energy storage. The proteomic analysis revealed that “Ca. B. sinica” synthesized glycogen via three known glycogen biosynthesis pathways and simultaneously degraded during the progress of active anammox, implying that glycogen is being continuously recycled. When cells were starved, a part of stored glycogen was converted to trehalose, a potential stress protectant. This suggests that glycogen serves at least as a primary carbon source of trehalose synthesis for survival. This study provides the first physiological evidence of glycogen metabolism in anammox bacteria and its significance in survival under natural substrate-limited habitat.
Similar content being viewed by others
Introduction
Many microorganisms including chemolithoautotrophs synthesize and accumulate carbon and energy reserves such as polysaccharide (glycogen) to cope with starvation conditions. Glycogen is a major intracellular reserve polymer consisting of α-1,4-linked glucose subunits with α-1,6-linked glucose at the branching points [1]. Bacterial glycogen can be synthesized via three known pathways: the GlgC–GlgA pathway, the Rv3032 pathway and the GlgE pathway. Each glycogen biosynthesis pathway was briefly described in Supplementary information text and reviewed in detail by Chandra et al. [2]. On the other hand, glycogen is degraded to glucose-1-phosphate by debranching enzyme (GlgX) and glycogen phosphorylase (GlgP) or to trehalose via the (TreX)–TreY–TreZ pathway [2]. Furthermore, the glycogen metabolism is known to be closely interlinked with biosynthesis of the disaccharide trehalose [2,3,4].
The time courses of glycogen accumulation and degradation during cultivation had been studied in several microorganisms. Several bacterial species optimally synthesize glycogen during exponential growth and/or stationary phase, and thus glycogen synthesis is regarded as an indicator of entering the stationary phase [3, 5]. For example, chemolithoautotrophs such as an aerobic ammonium-oxidizing bacterium (AOB), Nitrosomonas europaea, and a methane-oxidizing bacterium, “Ca. Methylacidiphilum fumariolicum”, were able to fix CO2 and accumulate glycogen during stationary phase when nitrogen was limited and utilize glycogen as energy source for cell survival during starvation [6, 7]. Furthermore, it was reported that glycogen was constantly recycled during early exponential-growth phase (meaning synthesis and degradation occurred simultaneously) in Corynebacterium glutamicum and utilized as a carbon source for trehalose synthesis under hyperosmotic conditions [3]. In general, glycogen accumulation was significantly enhanced in most of the bacteria when nitrogen is limited, but carbon sources are present in excess.
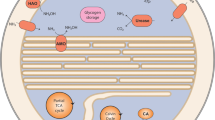
“Ca. Brocadia sinica” is an anaerobic ammonium-oxidizing (anammox) bacterium that can oxidize NH4+ with NO2− as an electron acceptor and fix CO2 into biomass via the Wood–Ljungdahl pathway [8, 9]. Studies so far suggested that anammox reaction proceeds by the following three steps with two unique intermediates such as nitric oxide (NO) and hydrazine (N2H4) [8, 9]. NO2− is initially reduced to NO by either cytochrome cd1-type or copper-containing nitrite reductase (NirS and NirK, respectively). However, “Ca. B. sinica” does not possess neither NirS nor NirK, which probably suggests the presence of the as-yet unidentified nitrite reductase [10]. The produced NO is then condensed with NH4+ to form N2H4 by hydrazine synthase (Hzs). The produced N2H4 is finally oxidized to N2 by hydrazine dehydrogenase (Hdh). Diverse anammox bacteria have been detected in various natural and man-made ecosystems [11, 12]. Anammox process has been used as a cost-effective and environment-friendly nitrogen removal process from wastewater [13, 14]. They are also responsible for a substantial part of the fixed nitrogen loss and re-oxidation of nitrite to nitrate in anoxic freshwater and marine ecosystems [15,16,17,18,19,20]. Despite of the engineering and ecological importance, pure cultures of anammox bacteria are not available so far. Five candidatus anammox bacteria genera (Brocadia, Kuenenia, Anammoxoglobus, Jettenia, and Scalindua) have been enriched from various samples and used for studying their ecophysiology and biochemistry [21].
There is evidence that anammox bacteria (Brocadia, Kuenenia, Anammoxoglobus, and Scalindua) could accumulate glycogen in their cytoplasmic compartment [22]. Furthermore, essential genes involved in the central carbon metabolic pathway including the classical glycogen biosynthesis (the GlgC–GlgA pathway) and degradation (the GlgX–GlgP pathway) were identified in the recently completed genomes of these anammox bacteria [23, 24]. However, very little is known about glycogen metabolism and the exact function in anammox bacteria. The previous study has suggested that glycogen could be utilized as an energy and carbon source under starvation and/or osmotic stress conditions [22]. However, this hypothesis needs to be verified experimentally because there were no significant differences in accumulation levels of intracellular glycogen in fresh and starved “Ca. Brocadia”-dominated biomass [25] and in anaerobically or anoxically starved “Ca. Kuenenia stuttgartiensis”-dominated granular biomass [26].
In the present study, “Ca. B. sinica“ was continuously cultured in a membrane bioreactor (MBR) with complete biomass retention, in which actively growing, near-zero growth, and starved phases were created by varying the nitrogen loading rate (NLR). The time course of intracellular glycogen accumulation and degradation in “Ca. B. sinica” was measured during different growth phases, which was linked to proteomic analysis to elucidate the glycogen metabolic pathways. Based on physiological studies, proteomic analysis, and electron microscopy, this study presents that the glycogen could be simultaneously synthesized and consumed (that is, constantly recycled) in “Ca. B. sinica” during growing and near-zero growth phase and that it was utilized at least as an energy and carbon source for trehalose biosynthesis under starvation conditions.
Materials and methods
Continuous culture
Planktonic enriched “Ca. Brocadia sinica” cells were continuously cultured at 37 °C in MBRs with complete biomass retention (alternatively called retentostats) as previously described (Fig. S1A–C) [27, 28]. All cells were in single cell state without presence of aggregates (Fig. S1D). The percentage of the anammox bacteria in the enriched culture was more than 96% as determined by fluorescence in situ hybridization (FISH) analysis with AMX820 and EUB mix oligonucleotide probes [29, 30]. The measurements of 16S rRNA gene copy numbers with quantitative PCR revealed that “Ca. B. sinica” accounted for more than 99% of the anammox population [31]. The MBRs consisted of a membrane unit composed of 300 polyethylene hollow-fiber membrane tubes (pore size, 0.1 μm; tube diameter, 1 mm; length, 70 mm, Mitsubishi Rayon, Tokyo, Japan) as described previously [28, 31]. A constant volume (1.5 L) of culture medium was maintained with a water level sensor HL-S1A (ASONE, Osaka, Japan) and a peristaltic pump (MP-1000, EYELA, Tokyo, Japan) that was directly connected to the membrane unit. The biomass culture was continuously stirred at 200 rpm with a magnetic stirrer to keep in planktonic state. A mixed Ar and CO2 (95:5) gas was continuously supplied to the culture medium at a flow rate of 10 mL min−1 to avoid oxygen contamination. pH was not controlled but ranged between 7.0 and 8.0. Biomass culture (15 mL) was withdrawn every day to maintain the sludge retention time (SRT) at ~100 days.
The MBR was initially inoculated with “Ca. B. sinica” cells that were starved for NH4+ and NO2− for one week. The initial biomass concentration was adjusted to be ca. 0.05 mg-protein mL−1. Inorganic anammox-specific medium was continuously fed into the MBR. The inorganic medium contained (mg L−1): (NH4)2SO4 (2–10 mM), NaNO2 (2–10 mM), FeSO4·7H2O (9.0), EDTA·2Na (5.0), NaCl (1.0), KCl (1.4), CaCl2·2H2O (1.4), MgSO4·7H2O (1.0), NaHCO3 (84), KH2PO4 [32], and 0.5 mL L−1 of trace element solution II [33]. The feed medium was purged with nitrogen gas (99.99%) for 1 h to maintain anoxic condition. A N2 gas bag was connected to the medium tank to prevent oxygen contamination during the continuous operation [34]. The nitrogen loading rate (NLR) was increased by increasing the influent NH4+ and NO2− concentrations (from 2 to 10 mM) to promote bacterial growth. The hydraulic retention time (HRT) was fixed at 1 h. After reaching a certain biomass concentration (i.e., near-zero specific growth rate), the biomass was starved for NH4+ and NO2− by stopping the medium feed. The continuous MBR culture experiment was conducted in triplicate.
Chemical analyses
Influent and effluent (permeate) samples were collected on daily basis for measurements of the concentrations of NH4+, NO2−, and NO3−, respectively. Ammonium (NH4+) concentration was measured using the colourimetric method with the indophenol and hypochlorous acid [35]. The produced colour was measured by a plate reader ARVO-MX (Perkin Elmer Japan, Yokohama, Japan) at 600 nm wavelength. Nitrite (NO2−) concentration was measured using the N-(1-Naphthyl)ethylene diamine method, and the resulting pink azo dye colour was measured by the plate reader at 520 nm wavelength [36]. Nitrate (NO3−) concentration was quantified using an ion chromatograph IC-2010 (TOSOH, Tokyo, Japan) with 3.8 mM of NaHCO3 and 3 mM of Na2CO3 as an anion eluent. All samples were filtered by 0.2-μm-pore size membranes (Advantec Co., Ltd., Tokyo, Japan) prior to the measurement.
Biomass dry weight
Biomass culture (15 mL) was collected and centrifuged at 10,000×g for 5 min. The cell pellet was washed, resuspended in 2 mL of Milli-Q water and then transferred to 5 mL vials whose weight is already known. The vials were then centrifuged at 10,000×g for 5 min and the supernatant was carefully discarded. The remaining biomass pellet was freeze-dried for at least one day using a freeze drier FZ-2.5 (Labconco, Kansas City, MO, USA). The vials with dry biomass were weighted again, and the biomass dry weight was calculated as difference between the vial weight with and without dry biomass. The obtained dry biomass was also subjected to the following measurements of intracellular glycogen and trehalose contents.
Estimation of specific growth rate
The specific growth rate (μ) of “Ca. B. sinica” growing in the MBR (retentostat) was estimated with the following equation [37]:
where Cx is biomass concentration in the MBR (g L−1). The biomass accumulation rate (dCx/dt) was determined by fitting the measured biomass concentrations with the equation Cx = A eB·t + C as previously reported [37]. This fitting was performed using R software (R Foundation for Statistical Computing, Vienna) and minimizing the sum of squares of errors by varying A, B, and C.
Glycogen and trehalose assays
The intracellular glycogen content was determined by a two-step enzymatic assay as previously described [38]. In brief, the freeze-dried biomass was resuspended in 0.25 mL of 0.25 M Na2CO3 solution in a screw-top Eppendorf tube and incubated at 95 °C for 4 h. The cell suspension was brought to pH 5.2 by addition of 0.15 mL of 1 M acetic acid and 0.6 mL of 0.2 M Na-acetate (pH 5.2) and then divided into two subsamples for determination of intracellular glycogen and trehalose. One-half was incubated with Aspergillus niger amyloglucosidase (1.2 units mL−1) (Sigma-Aldrich, St. Louis, MD, USA) over night at 57 °C under constant agitation to convert glycogen into glucose. After the amyloglucosidase treatment, the cell suspensions were centrifuged at 5000×g for 3 min to remove the cell debris, and the resulting supernatant was adequately diluted and then subjected to glucose measurement. The negative control without the amyloglucosidase treatment was prepared for glycogen measurement in parallel. Glucose was quantified by the glucose oxidase kit (Sigma-Aldrich), and the absorbance was recorded at 520 nm by a plate reader ARVO MX (Perkin Elmer Japan, Yokohama, Japan) [7]. For trehalose measurement, the second half of cell suspension was centrifuged at 5000×g for 3 min, and trehalose concentration in the supernatant was directly quantified by using a commercially available trehalose assay kit according to the manufacturer’s protocol (Megazyme, Wicklow, Ireland) [39]. Protein concentration of each sample was also determined using the Lowry method [40].
Proteomic analysis
“Ca. B. sinica” biomass cultures were collected from the MBR in the growing (day 18), near-zero growth (day 31), and starved (day 50) phase, respectively (Experimental Run-2 in Supplementary Information). Approximately 80 mg of wet biomass was suspended in the buffer A containing 3-[(3-cholamidopropyl) dimethylammonio] propanesulfonate (CHAPS) (4%, w/v), Triton X-100 (2%, w/v), urea (7 M), thiourea (2 M), and dithiothreitol (60 mM). After 1 h of incubation on ice and centrifugation at 25,000×g for 1 h at 4 °C, the supernatant was recovered as soluble protein fraction A. The remaining biomass pellet was resuspended in the buffer B containing SDS (4%, w/v), glycerol (20%, v/v), Tris–HCl (0.125 M, pH 8.4) and dithiothreitol (20 mM). After boiling for 15 min and centrifugation at 25,000×g for 1 h at 20 °C, the supernatant was recovered as soluble protein fraction B. Proteins in the soluble protein fractions A and B were loaded onto SDS containing polyacrylamide gels (NuPAGE 4–12% Bis–Tris gel) (Life Technologies, Carlsbad, CA, USA), respectively, and stained with Simplyblue staining solution (Life Technologies) according to the manufacturer’s instructions. Gel lanes (7 cm in height) were cut into 10 gel slices, which were de-stained with acetonitrile and subjected to nano-LC/MS/MS analysis after in-gel tryptic digestion [41]. Peptide mass fingerprints were analysed using the MASCOT search programme version 2.3.01 [42]. The amino acid sequences of protein-coding genes located in “Ca. B. sinica” genome (GenBank accession number; BAFH0100000–BAFH01000004) [43] were used as the reference database. Calculations of the exponentially modified protein abundance index (emPAI) and protein content index (PCI, in mol%) were conducted as previously described [44]. The sums of the PCI expressions of two protein fractions (A and B) were used in this study. The predicted genes sequences were used to search against the Clusters of Orthologous Genes (COG) and the Kyoto Encyclopaedia of Genes and Genomes (KEGG) databases using DIAMOND BLASTX with the e-value cutoff of 1e−5.
Transmission electron microscopy (TEM)
Sample preparation, staining, and electron microscopy were carried out as previously described by van Niftrik et al. [22]. Briefly, “Ca. B. sinica” cells were collected from the MBR, cryofixed and freeze-substituted using an EM PACT2 high pressure freezer (Leica Microsystems, Tokyo, Japan) and EM AFS2 freeze substitution processor (Leica Microsystems), respectively. Freeze substation with an acetone containing 2% (w/v) OsO4 (Nisshin EM, Tokyo, Japan) was carried out as follows: –90 °C for 50 h, increased to –60 °C with the increment of 2 °C per hour, kept at –60 °C for 8 h, and then increased to –30 °C with the increment of 2 °C per hour. The specimen was washed three times using acetone for 45 min at 25 °C, and then embedded into an Epon812 resin (Nisshin EM, Tokyo, Japan). The embedding and the subsequent polymerization was carried out as follows: kept in a 25% (v/v) Epon resin (diluted with acetone) at 25 °C for 16 h, 50% Epon resin for 24 h, 75% Epon resin for 24 h, 100% Epon resin at 35 °C for 7 h, 100% Epon resin at 30 °C for 17 h, and then polymerized at 60 °C for 3 days. Ultrathin sections (80–90 nm) were cut using a ULTRACUT S (Leica Microsystems, Tokyo, Japan). The sections were collected on Formvar/Carbon film-coated 100-mesh hexagonal golden grids and stained with 5-folds diluted TI blue solution (Nisshin EM) and lead citrate [45]. Electron microscopy was performed using a transmission electron microscope JEM-1200EX (JEOL, Tokyo, Japan) equipped with a TEM CCD camera Veleta (Olympus, Tokyo, Japan) at 80 kV [46].
For polysaccharide staining, the method of van Niftrik et al. [22] was slightly modified. The ultrathin sections were stained by 1% periodic acid (Sigma-Aldrich Japan, Tokyo, Japan) at 25 °C for 30 min and rinsed with Milli-Q water at 25 °C for 60 min. The sections were immersed into a 0.2% (w/v) thiocarbohydrazide (Sigma-Aldrich Japan) (diluted with 20% v/v acetone) and kept at 4 °C for 12 h. The sections were washed two times with 10% (v/v) acetic acid at 25 °C for 15 min, 2 times with 5% acetic acid for 5 min, with 2% acetic acid for 5 min, and then with Milli-Q water for 15 min. The washed sections were stained with 1% (w/v) silver albumin (Fuji-film Wako, Osaka, Japan) solution at 25 °C for 30 min, and washed with Milli-Q water for 15 min. Finally, the sections were immersed in a 5% (w/v) sodium thiosulfate (Sigma-Aldrich) and washed with Milli-Q water for 15 min. For this staining, the 1% periodic acid breaks up the polysaccharide (glycogen) molecules, to which thiocarbohydrazide can bind. The resulting combined molecules can be stained with silver albumin [22]. Therefore, the specimen without 1% periodic acid treatment was used as negative controls for glycogen staining in the present study.
Adenosine triphosphate (ATP)/adenosine diphosphate (ADP) measurement
ATP and ADP were quantified to determine the cellular energy charge. ATP was extracted using a mixture of 0.51 M tricarboxylic acid (TCA) and 100 mM EDTA for 30 min as previously described [47, 48]. The remaining TCA in the sample was removed by addition of water-saturated diethyl ether (cold) and purged with N2 gas for 5 min to solubilize the ether. ADP was first enzymatically converted to ATP by phosphoenolpyruvate (PEP) and pyruvate kinase (PK) [49, 50]. The conversion was conducted in the PEP-buffer containing 0.1 M Tris–EDTA buffer, 0.3 mM PEP, 9 mM MgCl2, 5 mM KCl, and PK (20 units mL−1). The total ATP and the ATP resulted from the ADP conversion were determined by using the firefly luciferase-based ATP measurement kit according to the manufacturer’s protocol (Kikkoman Biochemifa, Tokyo, Japan).
Results and discussion
Growth of “Ca. B. sinica” in MBR
One week starved planktonic “Ca. B. sinica” cells (made up >96% of population based on FISH analysis) were inoculated and continuously cultured for more than 2 months in a MBR with complete biomass retention (Fig. S1A–S1D). NLR was step-wisely increased from 0.05 to 0.27 kg-N m−3 day−1 by increasing the influent NH4+ and NO2− concentrations to promote the growth of “Ca. B. sinica” (Fig. 1A). After day 37, the NLR was fixed at ca. 0.27 kg-N m−3 day−1 and kept for 22 days. Thereafter the substrate feeding was completely stopped at day 58. The effluent NO2− concentration was near zero throughout the experiment, while effluent ammonium was varying between 1.0 and 20.0 mg L−1 (Fig. 1B). The stoichiometric ratio of consumed NO2− and consumed NH4+ was 1.32 ± 0.23, which was close to the previously reported value [51], indicating anammox reaction was predominantly responsible for nitrogen conversion.
Time course of nitrogen (NH4+ and NO2−) loading rate (NLR) and removal rate (NRR) (A) and the concentrations of NH4+, NO2−, and NO3− in MBR culture medium (B) during 70 days of continuous operation. Blue, white, and green area indicate the growing phase, near-zero growth phase, and starved phase, respectively. The error bars show SD of duplicate measurement.
The growth of “Ca. B. sinica” was assessed by measuring the protein concentration (Fig. 2). The protein concentration steadily increased to 0.26 mg-BSA mL−1 at day 37 and thereafter decreased slightly due to a limited substrate supply. After the substrate feeding was completely stopped at 58 days, the protein concentration decreased drastically. Time course of the specific growth rate (µ) was estimated from the measured protein concentration (Fig. 2). The estimated µ asymptotically decreased from 0.19 d−1, which was about half of µmax of “Ca. B. sinica” [31], at the beginning of operation to 0.0023 d−1 at day 37. Thereafter the specific growth rate (µ) reached near-zero growth rates (the net growth rate defined as µ - decay rate became negative). According to the estimated µ profile, the growth of “Ca. B. sinica” was classified into the growing phase (0–37 days), near-zero growth phase (38–58 days), and starved phase (59–70 days), respectively (Fig. 2).
The line indicates the specific growth rate (μ) that was estimated by fitting the measured protein concentrations with the equation Cx = A·eB·t + C as previously reported [37]. Blue, white, and green area indicate the growing phase, near-zero growth phase, and starved phase, respectively. The error bars show SD of duplicate measurement.
Intracellular glycogen metabolism
Intracellular glycogen content was very low at the beginning of cultivation, which was probably because one-week starved cells were inoculated (Fig. 3). However, intracellular glycogen content steadily increased and reached 32.6 mg-glucose (g-dry biomass wt)−1 as cells approaching to near-zero growth, which might indicate that substrate limitation might enhance glycogen accumulation. Thereafter, the glycogen content begun to decrease in the near-zero growth phase, during which the protein concentration also gradually decreased due to the limited substrate supply (the NLR was fixed at 0.27 kg-N m−3 day−1). When the cells were starved by completely ceasing the substrate supply, the glycogen content further decreased rapidly. Intriguingly, glycogen accumulation was immediately observed when the substrate supply was resumed after 16-day starvation (Figs. S2 and S3). Reproducible trends in glycogen accumulation and consumption were observed in independent triplicated MBR runs. Thus, the results of the third run were shown in the main text, whereas ones of the first and second run can be seen in the Supplementary Information Figs. S2–S7.
However, this growth phase-dependent phenomenon has not been observed in previous studies. No significant changes in intracellular glycogen levels were observed in the starved “Ca. Brocadia”-dominated [25] and “Ca. K. stuttgartiensis”-dominated granular biomass [26]. The intracellular glycogen levels of planktonic “Ca. B. sinica” cells observed in the present study were lower than those found in the “Ca. K. stuttgartiensis” granular biomass (6.55–8.48 mmol glucose g-VSS−1) [26]. Since it has been reported that glycogen stores were used for biofilm formation [52], it would be interesting to further investigate the effect of granule or biofilm formation on glycogen storage or vice versa.
“Ca. B. sinica” cells taken from the growing, near-zero growth, and starved phase were stained with 1% silver albumin (polysaccharide staining) with and without 1% periodic acid treatment and observed by TEM (Fig. 4). Electron dense particles were detected in the cytoplasmic compartment of 1% periodic acid-treated cells. The actively growing cells contained few and small dense particles, whereas more abundant and larger electron dense particles were detected in the cells taken from the near-zero growth phase. These TEM images corresponded to the intracellular glycogen content profile (Fig. 3). The previous TEM study showed that “Ca. K. stuttgartiensis” cells specially seem to store larger amounts of glycogen than other anammox genera including “Ca. Brocadia fulgida” [22], which is consist with the higher glycogen contents found in the starved “Ca. K. stuttgartiensis” biomass [26]. The difference in glycogen storage capacity among anammox bacteria species should be investigated in detail, if any.
Transmission electron micrographs showing glycogen accumulation in the cytoplasmic compartment of “Ca. B. sinica” cells in growing (A), near-zero growth (B and C), and starved (D) phase, respectively. Negative control incubated with water instead of periodic acid (E) showed no glycogen staining. The scale bars in panel A–D are 200 nm, and the one in panel E is 500 nm.
Intracellular glycogen content sharply decreased from 20.5 mg-glucose (g-biomass dry wt)−1 to <3 mg-glucose (g-biomass dry wt)−1 and concomitantly trehalose accumulated up to 1.0 mg-trehalose (g-biomass dry wt)−1 immediately after the substrate feed was stopped at day 58, suggesting that a part of glycogen was converted to trehalose (Fig. 5). However, since the amount of produced trehalose was only about 10% of the consumed glycogen, other metabolites of glycogen degradation could be present or used for energy generation. Trehalose is known to be accumulated in bacterial cells as a response to a variety of stresses such as heat, dry, starvation, and osmotic stress [4, 53]. The produced trehalose decreased rapidly to the original level (<0.2 mg-trehalose (g-biomass dry wt)−1) within a week. Probably, trehalose is broken down to glucose, which is eventually used as an energy and/or carbon source. Trehalose accumulation in anammox bacteria under substrate-limited conditions was reported for the first time. The degradation of trehalose is not well understood and thus needs further investigation.
Intracellular energy contents (ATP/ADP ratio)
The intracellular pools of ATP and ADP play an important role in the regulation of gluconeogenesis and glycolysis [32]. In general, glycolysis (glycogen degradation, energy generating process) is promoted when the ATP/ADP ratio is low, whereas the gluconeogenesis (toward glycogen synthesis, energy demanding process) is activated when the ATP/ADP ratio is high [54]. Thus, the ATP/ADP ratio seems to regulate the carbon flow in bacteria, which is entirely dependent on growth condition.
A key regulatory (rate-limiting) step of glycogen biosynthesis pathway is catalysed by glucose-1-phosphate adenylyltransferase (GlgC; EC 2.7.7.27); ATP + glucose-1-phosphate (Glc-1P) ⇆ ADP-Glc + PPi). This enzymatic reaction is allosterically regulated by intracellular energy level, and ATP generation negatively affects glycogen accumulation [6, 55,56,57]. Therefore, the ratio of ATP to ADP was measured during cultivation to correlate to the glycogen accumulation (Fig. 3). The ATP/ADP ratio steadily increased from the onset of cultivation but sharply declined after day 18. Thereafter, the intracellular glycogen content increased with decreasing the ATP/ADP ratio, indicating that excess ATP diverted toward glycogen biosynthesis. The stored glycogen begun to be utilized gradually at onset of near-zero growth phase and further decreased rapidly during the starved phase (no substrate feeding). The ATP/ADP ratio remained at low levels in the near-zero growth phase and further decreased in the starved phase. The reproducibility of this phenomenon was confirmed by a replicated experimental run (Fig. S6).
Proteomic analysis
Overall
A total of 1550, 1420, and 1276 proteins were identified in the growing, near-zero growth, and starved cells, respectively, with using more than 1 unique peptide-filtering criteria [44], representing 39.6–32.6% of all predicted proteins encoded in the “Ca. B. sinica” genome (Table S1). The growing and near-zero growth cells shared 910 proteins, the growing and starved cells shared 807 proteins, the near-zero growth and starved cells shared 761 proteins (Fig. S8). Approximately 70% of the identified proteins were associated with the available groups in COG classification, while the remaining proteins were belonged to the group COG R/S, ‘no hit’, ‘no related COG’ which indicated the hypothetical and poorly characterized proteins.
Most of the proteins that most highly expressed in growing, near-zero growth, and starved cells were associated with energy production and conversion (COG C) (Table S2). The second-high abundance proteins in growing and near-zero growth cells were associated with the post translational modification, protein turnover, and chaperones (COG O), whereas those in starved cells were involved in translation, ribosomal structure, and biogenesis (COG J). The proteins associated with amino acid transportation (COG E) were expressed higher in growing and near-zero growth cells, whereas heat shock proteins, ribosomal proteins, proteins associated with cell motility and chaperones were highly expressed under starved condition. It should be noted that many hypothetical proteins with unknown function that do not appear in the KEGG database were also expressed in high levels under the starved condition (Table S3). Thus, it is currently difficult to evaluate if starvation or near-zero growth conditions significantly affect metabolisms or pathways other than the nitrogen and carbon metabolisms.
The expression of proteins potentially involved in anammox reaction and central carbon metabolic pathways (i.e., glycogen and trehalose metabolism, Wood–Ljungdahl pathway, (reductive) TCA cycle, pentose phosphate pathway, and glycolysis/glyconeogenesis pathway) were further analysed for “Ca. B. sinica” cells in the growing, near-zero growth, and starved phase, respectively. The relative abundance represented as Protein Content Index (PCI, in mol%) values of the respective enzymes involved in anammox and central carbon metabolic pathways in each growth phase were summarized in Tables S4–S9.
Anammox metabolism
The expression levels (i.e., the PCI values (mol %)) of anammox-related enzymes were two orders of magnitude higher than those of the central carbon metabolic pathways. Nitrite oxidoreductase (Nxr), hydrazine oxidoreductase (Hzs), and hydrazine dehydrogenase (Hdh) were highly expressed in all growth phases even in starved phase (Table S4). This indicates that these enzymes were constitutively expressed and considerably stable even under the starved condition. These results suggest that chemolithoautotrophic “Ca. B. sinica” does not appear to shut down all these anammox-related (energy-generating) enzymes under starved conditions, which allows generating energy as soon as substrates becomes available. It was demonstrated that restarting substrate feeding resulted in immediate recovery of anammox activity after 16-day starvation in the present study (Fig. S2A). Similar phenomenon has been reported for autotrophic aerobic ammonium-oxidizing bacteria (AOB) [58]. Nitrosomonas could maintain high abundance and activity of ammonia monooxygenase (AMO) and hydroxylamine oxidoreductase (HAO) over prolonged starvation periods, while their transcriptions completely ceased shortly after the depletion of ammonia [59, 60]. However, it is unclear how they could maintain these enzymes active for a long time period without energy substrates.
Central carbon metabolism
Based on the genomic analysis and previous studies, it has been proposed that “Ca. B. sinica” assimilates CO2 via the Wood–Ljungdahl pathway using reducing power (equivalent) generated from nitrite oxidation to nitrate [8, 9, 23]. The relative expression levels of respective enzymes involved in the Wood–Ljungdahl pathway were represented as the PCI values (mol%) based on proteomic analysis (Fig. 6). The high abundances of key enzymes involved in the Wood–Ljungdahl pathway, such as CO dehydrogenase (CooS) and acetyl-CoA decarbonylase/synthase CODH/ACS complex (CdhDE) were detected in the growing and near-zero growth phase, whereas those expressions were very low in the starved phase (Table S5 and Fig. 6). This suggests that CO2 assimilation to acetyl-CoA is active in the growing and near-zero growth phase, reflecting the biomass growth profile (Fig. 2).
Protein expression level heatmaps of central carbon metabolic pathways (i.e., the Wood–Ljungdahl pathway, glycolysis pathway, and pentose phosphate pathway (reductive) tricarboxylic acid (TCA) cycle) in “Ca. B. sinica”, which was harvested in the growing, near-zero growth, and starved phase, respectively. The relative abundance of proteins represented as Protein Content Index (PCI, in mol%) values was portrayed by different colours in each pathway; the darker colour indicates the higher abundance and the lighter colour indicates the lower abundance. The names of enzymes, reactions, and corresponding PCI values are described in detail in the Supplementary information Tables S5–S8. n.d. not detected.
Gluconeogenesis is an essential pathway to synthesize glycogen in “Ca. B. sinica” because they are unable to use exogenous hexoses as carbon source for cell growth. Pyruvate ferredoxin oxidoreductase (PorABCD) and pyruvate orthophosphate dikinase (Ppdk) were highly expressed in all growth phases (Table S6 and Fig. 6), which catalyse the conversion of acetyl-CoA to PEP via pyruvate. The formed PEP flows into the gluconeogenesis pathway, in which high abundances of all enzymes involved in gluconeogenesis including fructose-1,6-bisphosphatase (FBP) were identified (Fig. 6). However, the enzyme in the final step of gluconeogenesis, glucose-6 phosphatase (G6Pas), was not identified, which could suggest that assimilated carbon compounds were diverted from glucose-6 phosphate to synthesis of glycogen and trehalose and/or the pentose phosphate pathway.
Most of the enzymes involved in the pentose phosphate pathway were abundantly detected in oxidative (generation of NADPH) and non-oxidative pathway (production of ribose 5-phosphate, which is a precursor for the synthesis of nucleotides) in the growing phase but were low or absent in the near-zero growth and starved phase (Table S7 and Fig. 6). This implies the active generation of NADPH and nucleotides during the growing phase and its shutdown during nongrowth energy-limited starvation conditions.
In terms of the TCA cycle, pyruvate:ferredoxin (Fd) oxidoreductase (porABCD; acetyl-CoA + CO2 + 2Fdred + 2H+ ↔ pyruvate + CoA + 2Fdox), 2-ketoglutarate:ferredoxin oxidoreductase (KorAB) and fumarate reductase (Frd), which are key enzymes of the reductive TCA cycle, were expressed in high abundance during the growing phase (Table S8 and Fig. 6). However, another key enzyme, ATP-citrate lyase (ACL, citrate + ATP + CoA → oxaloacetate + acetyl-CoA + ADP + Pi), is missing. Furthermore, the genes specific for the oxidative TCA cycle except for 2-ketoglutarate dehydrogenase complex (OGDC) were also detected in moderate abundance during growing and near-zero growth phase with glycogen accumulation.
Notably, metabolic pathways of key metabolites of gluconeogenesis and glycolysis such as PEP and pyruvate are versatile. For gluconeogenesis, PEP can be formed from oxaloacetate, which is catalysed by phosphoenolpyruvate carboxykinase (PEPCK) with ATP (oxaloacetate + ATP → PEP + ADP + CO2). The abundance of PEPCK was, however, not as high as other gluconeogenesis-related enzymes, suggesting that PEP is mainly synthesized from acetyl-CoA via pyruvate by porABCD and ppdk. These results reveal that acetyl-CoA is apparently a key intermediate for the gluconeogenesis in chemolithoautotrophic “Ca. B. sinica”, which is mainly synthesized through the Wood–Ljungdahl pathway. “Ca. B. sinica” also could generate energy and/or intermediates for cell growth through the oxidative TCA cycle with continuous input of metabolites from the Wood–Ljungdahl pathway and/or reductive TCA cycle, leading to more energy-efficient cell growth.
Furthermore, incomplete classical TCA bypass routes such as the glyoxylate shunt [61] and 4-aminobutyric acid (GABA) shunt [62, 63] were identified (Table S8 and Fig. 6). Isocitrate seems to be converted into succinate and glyoxylate by isocitrate lyase (ICL) instead of into 2-ketoglutarate, which bypasses two decarboxylation steps (isocitrate–2-ketoglutarate–succinyl-CoA) in the TCA cycle. This corresponds to the absence of OGDC. However, malate synthase (MS) which catalyses condensation of glyoxylate and acetyl-CoA to yield malate is missing in the proteomic analysis. The generated glyoxylate could be converted to glycerate-2P via glycerate by glycerate 2-kinase (gck), which was also highly expressed in all growth phase. The resulting glycerate-2P is a precursor of glucose-6P, and the consecutive formation of glucose-6P yields glycogen in the end. It has been proposed that the possible role of the glyoxylate shunt could be a protective response to oxidative stress [61].
In addition to the glyoxylate shunt, key enzymes in the GABA shunt, glutamate dehydrogenase (GdhA), glutamate decarboxylase (GadA), and GABA aminotransferase (puuE, EC:2.6.1.19) were identified, but succinic semialdehyde dehydrogenase (SAADH/SsaD) is missing. The GABA shunt could be also viewed as bypassing two decarboxylation steps of the TCA cycle, 2-ketoglutarate dehydrogenase (OGDC) and succinyl-CoA synthetase (sucCD), which compensated for the low activity of both enzymes (Fig. 6). Furthermore, the GABA shunt could regulate the nitrogen metabolite levels through glutamate and has stress-associated and signalling roles [63]. However, its regulation and biological significance in “Ca. B. sinica” remain largely unclear.
Glycogen metabolism
Genomic analysis revealed that “Ca. B. sinica” appears to possess three known pathways for bacterial glycogen synthesis: the GlgC–GlgA pathway, the GlgE pathway, and the Rv3032 pathway [2]. In the classical GlgC–GlgA pathway, the synthesis proceeds by three enzymatic reactions: adenosine diphosphate glucose (ADP-glucose, a monosaccharide nucleotide diphosphate) is generated from glucose 1-phosphate by the action of glucose-1-phosphate adenylyltransferase (GlgC), and then ADP-glucose is subsequently polymerized by glycogen synthase (GlgA), which generates linear α-1,4 glucans (Fig. 7). The linear glucans are further converted to glycogen by the α-1,4-glucan-branching enzyme (GlgB), which mediates formation of α-1,6-glucosidic linkages. In the GlgE pathway, the linear glucans are generated from maltose 1-phosphate (a disaccharide phosphate) by the action of maltosyltransferase (GlgE) [64, 65]. Maltose 1-phosphate is generated from trehalose by two enzyme actions: maltose α-d-glucosyltransferase/α-amylase (TreS) converts trehalose into maltose and a maltokinase (Pep2) phosphorylates the maltose using ATP. In the Rv3032 pathway, Rv3032 has α-1,4-glucosyltransferase activity and uses uridine diphosphate glucose (UDP-glucose) as the building block of α-1,4 glucans [66, 67]. Branches (α-1,6-glycosidic linkages) are further formed by the action of a branching enzyme Rv3031 [67].
Protein expression level heatmaps of main glycogen and trehalose metabolic pathways in “Ca. B. sinica”, which was harvested in the growing, near-zero growth, and starved phase, respectively. The relative abundance of proteins represented as Protein Content Index (PCI, in mol%) values was portrayed by different colours; the darker colour indicates the higher abundance and the lighter colour indicates the lower abundance. The names of enzymes, reactions, and corresponding PCI values are described in detail in the Supplementary information Table S9. n.d: not detected. Blue arrows represent biosynthetic pathways of glycogen, green arrows represent degradation pathways of glycogen, and orange arrows represent biosynthetic pathways of trehalose, respectively.
On the other hand, glycogen can be degraded to glucose 1-phosphate, a key intermediate for glucan reassembly, by the highly conserved glycogen phosphorylase (GlgP) with the assistance of the glycogen debranching enzymes (GlgX) and α-1,4-glucanotransferase (MalQ). Since GlgP can only cleave α-1,4-glycosidic linkages down to four glucose residues, the assistance of GlgX and MalQ is required to further breakdown the remaining glycogen with glucosyltransferase (transfers three glucose residues from the four-residue glycogen branch to another branch) and glucosidase (cleaves the remaining α-1,6-glycosidic linkages), which generates glucose and linear chains of α-1,4-glucans. Furthermore, debranching enzyme/isoamylase (ISA, EC:3.2.1.68) was detected in the present study. ISA has only α-1,6-glucosidase activity and thus generates maltodextrin. Maltose can be released from α-1,6-branched glycogen as the smallest sugar. ISA is similar to an archaeal glycogen-debranching enzyme (TreX, 74% homology) that has dual activities for α-1,4-glucanotransferase and α-1,6-glucosidase [68]. Therefore, glycogen could be also degraded to trehalose by an alternative route of (ISA)–TreY–TreZ or ISA–TreS. Trehalose can be utilized to reassemble glucans.
According to proteomic analysis, phosphoglucomutase (Pgm) is a key enzyme to provide a key intermediate, glucose-1 phosphate, for all routes to the biosynthesis of both α-1,4-glucan and trehalose, which was highly expressed in the growing and near-zero growth phase (Table S9 and Fig. 7). The key enzymes of glycogen synthesis including GlgC and GlgA were moderately expressed in the growing and near-zero growth phase. However, both the GlgA and Rv3032 were not expressed during the starved phase. Interestingly, the relatively high expression of the GlgE was identified only in the growing and starved phase, indicating the alternative route for glucan synthesis from trehalose via maltose was also active although the maltokinase (Pep2) is missing in the “Ca. B. sinica” genome. There are two potential sources of trehalose for the GlgE pathway: either via the de novo GalU–OtsA–OtsB route from activated glucose or via the (ISA)–TreY–TreZ route from α-1,6-branched phosphorylase-limit dextrin (Fig. 7). The details are described in the following trehalose metabolism section. The α-1,4-glucan-branching enzyme (GlgB), but not Rv3031, was moderately expressed in the growing and near-zero growth phase. The different biosynthetic routes of α-1,4-glucan could share the branching enzyme, which could support the notion that the GlgB-branching enzyme is more often associated with the GlgE pathway than with the GlgC–GlgA pathway [2].
On the other hand, the enzymes responsible for the glycogen degradation, starch phosphorylase (GlgP) and glycogen debranching enzymes (ISA) were moderately expressed during all growth phases. However, the most common debranching enzyme (GlgX) and α-1,4-glucanotransferase (MalQ) were sporadically expressed at low levels. In addition, AGL, which has dual activities for α-1,4-glucanotransferase and α-1,6-glucosidase, was detected at low levels only in near-zero growth phase. Furthermore, an essential enzyme of glycogen degradation, ADP-sugar-pyrophosphatase (AspP), which catalyses the conversion of ADP-glucose to glucose 1-phosphate, was only detected during the starved phase. AspP functions are also tightly regulated to prevent ADP-glucose accumulation and to divert the carbon flux from glycogen biosynthesis to other metabolic pathways in response to starvation [69].
Taken together, glycogen could be synthesized via three known pathways during the growing and near-zero growth phase; the GlgC–GlgA and the Rv3032 pathways from nucleotide sugars (ADP-glucose and UDP-glucose) and the GlgE pathway from a disaccharide intermediate (maltose-1P). Meanwhile, its degradation appears to be simultaneously occurring through the GlgP and TreYZ pathway, which generates key intermediates (i.e., glucose-1P and trehalose, respectively) for glucan reassembly. This indicates that a part of glycogen is being continuously recycled during the growing and near-zero growth phase, leading to accumulation of glycogen as a result of higher production rate. On the other hand, during the starved phase the glycogen production from nucleotide sugars (ADP- and UDP-glucose) appears to cease due to absence of GlgA and Rv3032, whereas GlgP- and TreYZ-mediated degradation (regeneration of Glucose-1P and trehalose) still continuously occurred. Thus, the intracellular glycogen content decreased rapidly (Fig. 3). The regenerated glucose-1 phosphate and trehalose could be used for other purposes.
Trehalose metabolism
Glycogen metabolism is closely interlinked to trehalose metabolism [2,3,4]. Trehalose is a stable compatible solute and stress protectant, which is composed of two glucose molecules connected through an α-1,1-glycosidic bond. Furthermore, trehalose can be used as an energy storage [70].
Five major pathways are known for trehalose biosynthesis in bacteria; from glycolytic intermediates like glucose-1 phosphate (a monosaccharide) via the GalU–OtsAB, GlgC–TreT, and TreP pathways, from maltose (a disaccharide) via the TreS pathway and from α-1,4-glucans (glycogen) via the TreYZ pathway (Fig. 7). In the GalU–OtsAB and GlgC–TreT pathways, nucleotide sugars (ADP-glucose and UDP-glucose) are used as building blocks. In the TreS pathway, TreS converts maltose directly into trehalose by converting α-1,4 glycosidic bond to α-1,1 glycosidic bond [71, 72]. In the TreYZ pathway, glycogen is first debranched into linear α-1,4-glucans (maltodextrin) with glycogen debranching enzymes (ISA, AGL, and/or GlgP with GlgX-MalQ). Maltodextrin is then converted to trehalose with glucosylmutase (TreY), which converts the terminal α-1,4-bond (reducing end) of a linear glucan into an α-1,1-bond, forming maltooligosyltrehalose, and the formed trehalose is released with trehalohydrolase (TreZ) [2, 3, 73].
Genomic analysis revealed that “Ca. B. sinica” possesses almost all the genes encoding enzymes involved in these trehalose biosynthesis pathways [28]. According to proteomic analysis, the enzymes involved in the TreP pathway from glucose-1 phosphate and the TreT pathway from ADP-glucose were highly expressed only in the growing phase, whereas the OtsAB pathway via UDP-glucose was less abundant in all growth phases. Furthermore, the UTP-glucose-1-phosphate uridylyltransferase (GalU), which forms UDP-glucose from glucose-1-phosphate, was not detected. The maltose pathway (TreS) was identified only in the growing and near-zero growth phase. On the other hand, the TreYZ pathway was detected at moderate levels in all growth phases.
Taken together, a key intermediate for all biosynthesis routes (i.e., glucose-1P) was sufficiently supplied from the gluconeogenesis in the growing phase and thus diverted toward trehalose biosynthesis via the de novo TreP and TreT pathways. In addition, trehalose was also produced from glycogen via the TreYZ pathway and from maltose via the TreS pathway in the growing and near-zero growth phase. By contrast, glycogen became a primary source of trehalose via the TreYZ pathway in the starved phase. Trehalose produced at onset of the starved phase was consumed rapidly (Fig. 5). These results imply that trehalose was utilized as a source of energy and could function as a stress protectant to protect proteins and cell membranes from denaturation or aggregation and other damages under stressed conditions. However, further study is required to understand the relationship between accumulation of glycogen and trehalose and stress resistance in anammox bacteria in detail.
Roles of glycogen metabolism in chemolithoautotrophic “Ca. B. sinica”
The ability of “Ca. B. sinica” to use reduced carbon compounds (e.g., formate, acetate, and propionate) as an energy and carbon source has been reported [8, 28]. In the present study, “Ca. B. sinica” was cultured under autotrophic growth conditions (without addition of those reduced carbon compounds) with CO2 as sole carbon source. The stored glycogen was started being utilized at the onset of entering the near-zero growth phase (Fig. 3). The stoichiometric ratio of produced NO3− and consumed NH4+ before glycogen utilization was 0.364 ± 0.165 and decreased to 0.205 ± 0.095 during glycogen utilization (a statistically significant difference; P < 0.015). This phenomenon was reproducible in a replicated run (0.355 ± 0.119 in the growing phase, 0.095 ± 0.079 in the near-zero growth phase, and 0.420 ± 0.147 in the second growing phase (P < 0.0001)) (Fig. S4). These results might suggest that glycogen degradation provides the cells with alternative reducing power (NADH and/or NADPH) and energy for CO2 fixation or metabolites of glycogen degradation (e.g., glucose) were directly utilized as precursors of biomass synthesis, resulting in a lower energy requirement for energetically expensive CO2 fixation.
Apparently, “Ca. B. sinica” possesses multiple biosynthesis pathways of both α-1,4-glucan and trehalose. In particular, the existence of three α-1,4-glucan biosynthesis pathways in “Ca. B. sinica” is revealed for the first time. In addition to the classical GlgC–GlgA pathway, the GlgE (although Pep2 is missing) and Rv3032 pathways are identified by the proteomic analysis. The complete GlgE pathway has been identified in only 14% of sequenced bacteria genomes, and the Rv3032 pathway is more likely restricted to Mycobacteria [2]. Disruption of either of the GlgE or the Rv3032 pathway has no effect on the growth of Mycobacterium tuberculosis, but blocking both is lethal [65]. This implies that the pathways are partially redundant at least. It is still not clear how much these pathways are inter-connected and how they are regulated, which requires further studies. In addition, it would be interesting to investigate whether the physico-chemical nature of the α-1,4-glucans produced via these different routes is same. Notably, the GlgE pathway (TreS–Pep2–GlgE–GlgB) coupled with the TreYZ pathway seems to be a complemental way of cycling glycogen and trehalose. Glycogen is the major storage carbohydrate in “Ca. B. sinica” (glycogen constituted ~2–3% of biomass dry weight whereas trehalose constituted only <0.1%, Fig. 5). The role and regulation of both cycling pathways in chemolithoautotrophic anammox bacteria requires further investigation.
Conclusions
During the growing phase, the intracellular glycogen content in “Ca. B. sinica” was observed to increase while the specific growth rate (μ) and the ATP/ADP ratio was decreased. The stored glycogen begun to be utilized gradually at onset of entering the near-zero growth phase and degraded rapidly when the cells were starved. This indicates that stored glycogen is used as an energy storage at least. Furthermore, proteome analysis suggested that “Ca. B. sinica” possesses multiple biosynthesis pathways of both α-1,4-glucan and trehalose and that glycogen seems to be continuously synthesized and degraded (implying the recycling of glycogen) in the growing phase. A part of stored glycogen was transiently converted to trehalose when the cells were starved, implying that glycogen is used as a carbon source of trehalose biosynthesis serving as a stress protectant.
Glycogen appears to play relevant roles in the survival of anammox bacteria in natural habitat, and glycogen metabolism is highly interlinked with multiple and important cellular processes. Further study is required to understand the regulation of glycogen and trehalose metabolism and their significance in survival under substrate starvation conditions and biofilm formation.
References
Preiss J. Bacterial glycogen inclusions: enzymology and regulation of synthesis. In: Shively JM, editor. Inclusions in prokatyotes. Springer: Heidelberg, Germany, 2006. p. 71–108.
Chandra G, Chater KF, Bronemann S. Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiology. 2011;157:1565–72.
Seibold GM, Eikmanns BJ. The glgX gene product of Corynebacterium glutamicum is required for glycogen degradation and for fast adaptation to hyperosmotic stress. Microbiology. 2007;153:2212–20.
Ruhal R, Kataria R, Choudhury B. Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation. Microb Biotechnol. 2013;6:493–502.
Iglesias AA, Preiss J. Bacterial glycogen and plant starch biosynthesis. Biochem Educ. 1992;20:196–203.
Machtey M, Kuhn ML, Flasch DA, Aleanzi M, Ballicora MA, Igesias AA. Insights into glycogen metabolism in chemolithoautotrophic bacteria from distinctive kinetic and regulatory properties of ADP-glucose pyrophosphorylase from Nitrosomonas europaea. J Bacteriol. 2012;194:6056–65.
Khadem AF, van Teeseling MCF, van Niftrik L, Jetten MSM, Op den Camp HJM, Pol A. Genomic and physiological analysis of carbon storage in the verrumicrobial methanotroph “Ca. Methylacidiphilum fumariolicum” SoIV. Front Microbiol 2012; 3: article 345.
Kartal B, Almeida NM, Maalcke WJ, Op den Camp HJM, Jetten MSM, Keltjens JT. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev. 2013;37:428–61.
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, et al. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–4.
Oshiki M, Ali M, Shinyako-Hata K, Satoh H, Okabe S. Hydroxylamine-dependent anaerobic ammonium oxidation (Anammox) by “Candidatus Brocadia sinica”. Environ Microbiol. 2016;18:3133–43.
Oshiki M, Satoh H, Okabe S. Ecology and physiology of anaerobic ammonium oxidizing (anammox) bacteria. Environ Microbiol. 2016;18:2784–96.
Sonthiphand P, Hall MW, Neufeld JD. Biogeography of anaerobic ammonia-oxidizing (anammox) bacteria. Front Microbiol. 2014;5:399.
Ali M, Okabe S. Anammox-based technologies for nitrogen removal: advances in process start-up and remaining issues. Chemosphere. 2015;141:144–53.
Kartal B, Kuenen JG, van Loosdrecht MC. Sewage treatment with anammox. Science. 2010;328:702–3.
Humbert S, Tarnawski S, Fromin N, Mallet MP, Aragno M, Zopfi J. Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J. 2010;4:450–4.
Kobayashi K, Makabe A, Yano M, Oshiki M, Kindaichi T, Casciotti K, et al. Dual nitrogen and oxygen isotope fractionation during anaerobic ammonium oxidation by anammox bacteria. ISME J. 2019;13:2426–36.
Kalvelage T, Lavik G, Lam P, Contreras S, Arteaga L, Löscher CR, et al. Nitrogen cycling driven by organic matter export in the South Pacific oxygen minimum zone. Nat Geosci. 2013;6:228–34.
Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2005;437:349–55.
Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, et al. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci USA. 2005;102:6478–83.
Thamdrup B, Jensen MM, Ulloa O, Farias L, Escribano R. Anaerobic ammonium oxidation in the oxygen-deficient waters off northern Chile. Limnol Ocean. 2006;51:2145–56.
Zhang L, Okabe S. Ecological niche differentiation among anammox bacteria. Water Res. 2020;171:115468.
van Niftrik L, Geerts WJC, van Donselaar EG, Humbel BM, Webb RI, Fuerst JA, et al. Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage, and localization of cytochrome c proteins. J Bacteriol. 2008;190:708–17.
Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, et al. Molecular mechanism of anaerobic ammonium oxidation. Nature. 2011;479:127–30.
Lawson CE, Wu S, Bhattacharjee AS, Hamilton JH, McMahon KD, Goel R, et al. Metabolic network analysis reveals microbial community interactions in anammox granules. Nat Commun. 2017;8:15416.
Carvajal-Arroyo JM, Puyol D, Li G, Swartwout A, Sierra-Alvarez R, Field JA. Starved anammox cells are less resistant to NO2− inhibition. Water Res. 2014;65:170–6.
Ma X, Wang Y. Anammox bacteria exhibit capacity to withstand long-term starvation stress: a proteomic-based investigation of survival mechanisms. Chemosphere. 2018;211:952–61.
Oshiki M, Shimokawa M, Fujii N, Satoh H, Okabe S. Physiological characteristics of the anaerobic ammonium-oxidizing bacterium ‘Candidatus Brocadia sinica’. Microbiology. 2011;157:1706–13.
Oshiki M, Awata T, Kindaichi T, Satoh H, Okabe S. Cultivation of planktonic anaerobic ammonium oxidation (anammox) bacteria using membrane bioreactor. Microbes Environ. 2013;28:436–43.
Kindaichi T, Tsushima I, Ogasawara Y, Shimokawa M, Ozaki N, Satoh H, et al. In situ activity and spatial organization of anaerobic ammonium-oxidizing (Anammox) bacteria in biofilms. Appl Environ Microbiol. 2007;73:4931–9.
Okabe S, Satoh H, Watanabe Y. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1999;65:3182–91.
Zhang L, Narita Y, Gao L, Ali M, Oshiki M, Okabe S. Maximum specific growth rate of anammox bacteria revisited. Water Res. 2017;116:296–303.
Kilstrup M, Hammer K, Jensen PR, Martinussen J. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol Rev. 2005;29:555e590.
van de Graaf AA, de Bruijn P, Robertson LA, Jetten MSM, Kuenen JG. Autotrophic growth of anaerobic ammonium-oxidizing microorganisms in a fluidized bed reactor. Microbiology. 1996;142:2187–96.
Tsushima I, Ogasawara Y, Kindaichi T, Satoh H, Okabe S. Development of high-rate anaerobic ammonium-oxidizing (anammox) biofilm reactors. Water Res. 2007;41:1623–34.
Solorzano L. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol Oceanogr. 1969;14:799–801.
Nagaraja P, Shivaswamy M, Kumar H. Highly sensitive N-(1-Naphthyl)ethylene diamine method for the spectrophotometric determination of trace amounts of nitrite in various water samples. Anal Chem 2001;80:39–48.
Overkamp W, Ercan O, Herber M, van Maris AJA, Kleerebezem M, Kuipers OP. Physiological and cell morphology adaptation of Bacillus subtilis at near-zero specific growth rates: a transcriptome analysis. Environ Microbiol. 2015;17:346–63.
Parrou JL, Francois J. A simplified procedure for a rapid and reliable assay of both glycogen and treahalose in whole yeast cells. Anal Biochem. 1997;248:186–8.
Neves MJ, Terenzi HF, Leone FA, Jorge JA. Quantification of trehalose in biological samples with a conidial trehalase from the thermophilic fungus Humicola grisea var. thermoidea. World J Microbiol Biotech. 1994;10:17–19.
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75.
Kasahara Y, Morimoto H, Kuwano M, Kadoya R. Genome-wide analytical approaches using semiquantitative expression proteomics for aromatic hydrocarbon metabolism in Pseudomonas putida F1. J Microbiol Methods. 2012;91:434–42.
Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67.
Oshiki M, Shinyako-Hata K, Satoh H, Okabe S. Draft genome sequence of an anaerobic ammonium-oxidizing bacterium, “Candidatus Brocadia sinica. Genome Announc. 2015;3:e00267–15.
Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteom. 2005;4:1265–72.
Reynold ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–12.
Ali M, Oshiki M, Awata T, Isobe K, Kimura Z, Yoshikawa H, et al. Physiological characterization of anaerobic ammonium oxidizing bacterium, “Ca. Jettenia caeni”. Environ Microbiol. 2014;7:2172–89.
Lundin A, Thore A. Comparison of methods for extraction of bacterial adenine nucleotides determined by firefly assay. Appl Microbiol. 1975;30:713–21.
Larsson CM, Olsson T. Firefly assay of adenine nucleotides from algae: comparison of extraction methods. Plant Cell Physiol. 1979;20:145–55.
Spielmann H, Jacob-Muller U, Schulz P. Simple assay of 0.1-1.0 pmol of ATP, ADP, and AMP in single somatic cells using purified luciferin luciferase. Anal Biochem. 1981;113:172–8.
Karl DM, Holm-Hansen O. Methodology and measurement of adenylate energy charge ratio in environmental samples. Mar Biol. 1978;48:185–97.
Lotti T, Kleerebezem R, Lubello C, van Loosdrecht MCM. Physiological and kinetic characterization of a suspended cell anammox culture. Water Res. 2014;60:1–14.
Bonafonte MA, Solano C, Sesma B, Alvarez M, Montuenga L, Garcia-Ros D, et al. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol Lett. 2000;191:31–36.
Argüelles JC. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch Microbiol. 2000;174:217–24.
Feng Y, Zhao Y, Guo Y, Liu S. Microbial transcript and metabolome analysis uncover discrepant metabolic pathways in autothropic and mixotrophic anammox consortia. Water Res. 2018;128:402–11.
Eydallin G, Viale AM, Moran-Zorzano MT, Munoz FJ, Montero M, Baroja-Fernandez E, et al. Genome-wide screening of genes affecting glycogen metabolism in Escherichia coli K-12. FEBS Lett. 2007;581:2947–53.
Preiss J. Glycogen: biosynthesis and regulation. EcoSal Plus 2014. https://doi.org/10.1128/ecosalplus.4.7.4.
Rodriguez-Lopez M, Baroja-Fernandez E, Zandueta-Criado A, Pozueta-Romero J. Adenosine diphosphate glucose pyrophosphatase: a plastidial phosphodiesterase that prevents starch biosynthesis. Proc Natl Acad Sci USA. 2000;97:8705–10.
Geets J, Boon N, Verstraete W. Startegies of aerobic ammonia-oxidizing bacteria for coping with nutrient and oxygen fluctuations. FEMS Microbiol Ecol. 2006;58:1–13.
Sayavedra-Soto L, Hommes N, Russell SA, Arp DJ. Induction of ammonia monooxygenase and hydroxylamine oxidoreductase mRNAs by ammonium in Nitrosomonas europaea. Mol Microbiol. 1996;20:541–8.
Wilhelm R, Abeliovich A, Nejidat A. Effect of long-term ammonia starvation on the oxidation of ammonia and hydrolylamine by Nitrosomonas europaea. J Biochem. 1998;124:811–5.
Ahn S, Jung J, In-Ae Jang I-A, Madsen EL, Park W. Role of glyoxylate shunt in oxidative stress response. J Biol Chem. 2016;291:11928–38.
Zhang S, Bryant DA. The tricarboxylic acid cycle in cyanobacteria. Science. 2011;334:1551–3.
Zhang S, Qian X, Chang S, Dismukes GC, Bryant DA. Natural and synthetic variants of the tricarboxylic acid cycle in cyanobacteria: introduction of the GABA shunt into Synechococcus sp. PCC 7002. Front Microbiol. 2016;7:1972.
Elbein AD, Pastuszak I, Tackett AJ, Wilson T, Pan YT. Last step in the conversion of trehalose to glycogen: a mycobacterial enzyme that transfers maltose from maltose 1-phosphate to glycogen. J Biol Chem. 2010;285:9803–12.
Kalscheuer R, Syson K, Veeraraghavan U, Weinrick B, Biermann KE, Liu Z, et al. Self-poisoning of Mycobacterium tuberculosis by targeting GlgE in an α-glucan pathway. Nat Chem Biol. 2010;6:376–84.
Kaur D, Pham H, Larrouy-Maumus G, Riviere M, Vissa V, Guerin ME, et al. Initiation of methylglucose lipopolysaccharide biosynthesis in mycobacteria. PLoS ONE. 2009;4:e5447.
Stadthagen G, Sambou T, Guerin M, Barilone N, Boudou F, Kordulakova J, et al. Genetic basis for the biosynthesis of methylglucose lipopolysaccharides in Mycobacterium tuberculosis. J Biol Chem. 2007;282:27270–6.
Woo EJ, Lee S, Cha H, Park JT, Yoon SM, Song HN, et al. Structural insight into the bifunctional mechanism of the glycogen-debranching enzyme TreX from the Archaeon Sulfolobus solfataricus. J Biol Chem. 2008;283:28641–8.
Wilson WA, Roach PJ, Montero M, Baroja-Fernandez E, Munoz FJ, Eydallin G, et al. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev. 2010;34:952–85.
Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17R–27R.
Jiang L, Lin M, Zhang Y, Li Y, Xu X, Li S, et al. Identification and characterization of a novel trehalose synthase gene derived from saline–alkali soil metagenomes. PLoS ONE. 2013;8:e77437.
Wei YT, Zhu QX, Luo ZF, Lu FS, Chen FZ, Wang QY, et al. Cloning, expression and identification of a new trehalose synthase gene from Thermobifida fusca genome. Acta Biochim Biophys Sin. 2004;36:477–84.
Maruta K, Mitsuzumi H, Nakada T, Kubota M, Chaen H, Fukuda S, et al. Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim Biophys Acta. 1996;1291:177–81.
Acknowledgements
This research was financially supported by Nagase Science and Technology Foundation, Institute for Fermentation, Osaka (IFO), and JSPS KAKENHI (Grant Number 19H0077609), which were granted to SO. AAS expresses her utmost gratitude to the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Government of Japan for providing the scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Okabe, S., Shafdar, A.A., Kobayashi, K. et al. Glycogen metabolism of the anammox bacterium “Candidatus Brocadia sinica”. ISME J 15, 1287–1301 (2021). https://doi.org/10.1038/s41396-020-00850-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-020-00850-5
This article is cited by
-
Genome-resolved metagenomic analysis reveals different functional potentials of multiple Candidatus Brocadia species in a full-scale swine wastewater treatment system
Frontiers of Environmental Science & Engineering (2023)
-
Response and resilience of anammox consortia to nutrient starvation
Microbiome (2022)