Abstract
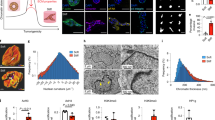
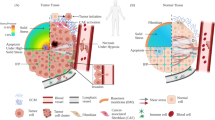
In many cancers, tumour progression is associated with increased tissue stiffness. Yet, the mechanisms associating tissue stiffness with tumorigenesis and malignant transformation are unclear. Here we show that in gastric cancer cells, the stiffness of the extracellular matrix reversibly regulates the DNA methylation of the promoter region of the mechanosensitive Yes-associated protein (YAP). Reciprocal interactions between YAP and the DNA methylation inhibitors GRHL2, TET2 and KMT2A can cause hypomethylation of the YAP promoter and stiffness-induced oncogenic activation of YAP. Direct alteration of extracellular cues via in situ matrix softening reversed YAP activity and the epigenetic program. Our findings suggest that epigenetic reprogramming of the mechanophysical properties of the extracellular microenvironment of solid tumours may represent a therapeutic strategy for the inhibition of cancer progression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data used to make the figures are available as Supplementary Information. Web links to publicly available transcriptomic datasets are provided in the Methods. All of the sequence data have been deposited to the NCBI Sequence Read Archive with the BioProject ID PRJNA673653 and SRP accession code SRP290642. Raw data are available from the corresponding authors upon reasonable request.
References
Crowder, S. W., Leonardo, V., Whittaker, T., Papathanasiou, P. & Stevens, M. M. Material cues as potent regulators of epigenetics and stem cell function. Cell Stem Cell 18, 39–52 (2016).
Chaudhuri, O. et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 13, 970–978 (2014).
Mohammadi, H. & Sahai, E. Mechanisms and impact of altered tumour mechanics. Nat. Cell Biol. 20, 766–774 (2018).
Humphrey, J. D., Dufresne, E. R. & Schwartz, M. A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 15, 802–812 (2014).
Malik, R., Lelkes, P. I. & Cukierman, E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 33, 230–236 (2015).
Lee, J. Y. et al. YAP-independent mechanotransduction drives breast cancer progression. Nat. Commun. 10, 1848 (2019).
Totaro, A., Panciera, T. & Piccolo, S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 20, 888–899 (2018).
Zanconato, F., Cordenonsi, M. & Piccolo, S. YAP and TAZ: a signalling hub of the tumour microenvironment. Nat. Rev. Cancer 19, 454–464 (2019).
Brusatin, G., Panciera, T., Gandin, A., Citron, A. & Piccolo, S. Biomaterials and engineered microenvironments to control YAP/TAZ-dependent cell behaviour. Nat. Mater. 17, 1063–1075 (2018).
Panciera, T., Azzolin, L., Cordenonsi, M. & Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 18, 758–770 (2017).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Hansen, C. G., Moroishi, T. & Guan, K. L. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 25, 499–513 (2015).
Zanconato, F., Cordenonsi, M. & Piccolo, S. YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803 (2016).
Schellenberg, A. et al. Matrix elasticity, replicative senescence and DNA methylation patterns of mesenchymal stem cells. Biomaterials 35, 6351–6358 (2014).
Xie, S. A. et al. Matrix stiffness determines the phenotype of vascular smooth muscle cell in vitro and in vivo: role of DNA methyltransferase 1. Biomaterials 155, 203–216 (2018).
Liu, Y. Y. et al. Fibrin stiffness mediates dormancy of tumor-repopulating cells via a Cdc42-driven Tet2 epigenetic program. Cancer Res. 78, 3926–3937 (2018).
Jang, M. et al. Increased extracellular matrix density disrupts E-cadherin/β-catenin complex in gastric cancer cells. Biomater. Sci. 2018, 2704–2713 (2018).
Zhou, Z. H. et al. Reorganized collagen in the tumor microenvironment of gastric cancer and its association with prognosis. J. Cancer 8, 1466–1476 (2017).
Lim, B. et al. Integrative genomics analysis reveals the multilevel dysregulation and oncogenic characteristics of TEAD4 in gastric cancer. Carcinogenesis 35, 1020–1027 (2014).
Shih, Y. L. et al. Quantitative methylation analysis reveals distinct association between PAX6 methylation and clinical characteristics with different viral infections in hepatocellular carcinoma. Clin. Epigenetics 8, 41 (2016).
Kang, M. H. et al. Verteporfin inhibits gastric cancer cell growth by suppressing adhesion molecule FAT1. Oncotarget 8, 98887–98897 (2017).
Yang, C., Tibbitt, M. W., Basta, L. & Anseth, K. S. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 13, 645–652 (2014).
Nasrollahi, S. et al. Past matrix stiffness primes epithelial cells and regulates their future collective migration through a mechanical memory. Biomaterials 146, 146–155 (2017).
Sun, Z., Guo, S. S. & Fassler, R. Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 (2016).
Nardone, G. et al. YAP regulates cell mechanics by controlling focal adhesion assembly. Nat. Commun. 8, 15321 (2017).
Werner, S. et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J. Biol. Chem. 288, 22993–23008 (2013).
Chen, W. et al. Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5′-CpG island in normal human keratinocytes. J. Biol. Chem. 285, 40852–40863 (2010).
Gontier, G. et al. Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 22, 1974–1981 (2018).
Huang, Y. C. et al. Epigenetic regulation of NOTCH1 and NOTCH3 by KMT2A inhibits glioma proliferation. Oncotarget 8, 63110–63120 (2017).
Chen, W. et al. Grainyhead-like 2 enhances the human telomerase reverse transcriptase gene expression by inhibiting DNA methylation at the 5′-CpG island in normal human keratinocytes. J. Biol. Chem. 285, 40852–40863 (2010).
Wang, L. et al. TET2 coactivates gene expression through demethylation of enhancers. Sci. Adv. 4, eaau6986 (2018).
Cierpicki, T. et al. Structure of the MLL CXXC domain–DNA complex and its functional role in MLL-AF9 leukemia. Nat. Struct. Mol. Biol. 17, 62–68 (2010).
Choi, W. et al. YAP/TAZ initiates gastric tumorigenesis via upregulation of MYC. Cancer Res. 78, 3306–3320 (2018).
Kang, W. et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin. Cancer Res. 17, 2130–2139 (2011).
Vining, K. H. & Mooney, D. J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 18, 728–742 (2017).
Albrengues, J. et al. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat. Commun. 6, 10204 (2015).
Stowers, R. S. et al. Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility. Nat. Biomed. Eng. 3, 1009–1019 (2019).
Ehrlich, M. DNA hypomethylation in cancer cells. Epigenomics 1, 239–259 (2009).
Yang, X. J., Lay, F., Han, H. & Jones, P. A. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol. Sci. b, 536–546 (2010).
Ramchandani, S., Bhattacharya, S. K., Cervoni, N. & Szyf, M. DNA methylation is a reversible biological signal. Proc. Natl Acad. Sci. USA 96, 6107–6112 (1999).
Kim, M. H. et al. Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. EMBO J. 35, 462–478 (2016).
Li, L. C. & Dahiya, R.MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 (2002).
Kuo, H. C. et al. DBCAT: database of CpG islands and analytical tools for identifying comprehensive methylation profiles in cancer cells. J. Comput. Biol. 18, 1013–1017 (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128 (2013).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. The molecular signatures database hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Oki, S. et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19, e46255 (2018).
Acknowledgements
This research was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2019R1A2C2084142 to P.K.), and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea (HI14C1324 to P.K. and J.-H.C.). This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation of Korea, funded by the Ministry of Science and ICT (NRF-2017M3A9A7050612 to J.K.C.).
Author information
Authors and Affiliations
Contributions
M.J. and P.K. designed the experiments. M.J. and S.W.O. performed the experiments and analysed the data. J.A., M.J. and J.K.C. performed the bioinformatics analysis. J.Y.L. and J.-H.C. helped with approval of the Institutional Review Board at the Yonsei University Severance Hospital and executed clinical applications. J.K. helped with the YAP depletion experiments. M.J., J.A., J.K.C., J.-H.C. and P.K. wrote the manuscript. All authors discussed the results and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Table 1.
Supplementary Data 1
Source data for the figures.
Supplementary Video 1
Snapshots of AGS cells in the stiff matrix, captured every 12 h over the course of 72 h.
Supplementary Video 2
Snapshots of AGS cells in the softened matrix, captured every 12 h over the course of 72 h.
Supplementary Video 3
Snapshots of AGS cells in the soft matrix, captured every 12 h over the course of 72 h.
Supplementary Video 4
Snapshots of AGS cells under control siRNA treatment in the stiff matrix, captured every 12 h over the course of 72 h.
Supplementary Video 5
Snapshots of AGS cells under control siRNA treatment in the soft matrix, captured every 12 h over the course of 72 h.
Supplementary Video 6
Snapshots of YAP-depleted AGS cells under siYAP treatment in the stiff matrix, captured every 12 h over the course of 72 h.
Supplementary Video 7
Snapshots of YAP-depleted AGS cells under siYAP treatment in the soft matrix, captured every 12 h over the course of 72 h.
Rights and permissions
About this article
Cite this article
Jang, M., An, J., Oh, S.W. et al. Matrix stiffness epigenetically regulates the oncogenic activation of the Yes-associated protein in gastric cancer. Nat Biomed Eng 5, 114–123 (2021). https://doi.org/10.1038/s41551-020-00657-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-020-00657-x
This article is cited by
-
Rebuilding the microenvironment of primary tumors in humans: a focus on stroma
Experimental & Molecular Medicine (2024)
-
Age-related matrix stiffening epigenetically regulates α-Klotho expression and compromises chondrocyte integrity
Nature Communications (2023)
-
Biophysics in tumor growth and progression: from single mechano-sensitive molecules to mechanomedicine
Oncogene (2023)
-
Cell–extracellular matrix mechanotransduction in 3D
Nature Reviews Molecular Cell Biology (2023)
-
A biomechanical view of epigenetic tumor regulation
Journal of Biological Physics (2023)