- 1USDA Forest Service, Forest Products Laboratory, Madison, WI, United States
- 2Department of Forestry and Wildlife Ecology, University of Wisconsin-Madison, Madison, WI, United States
Copper tolerance of brown-rot basidiomycete decay fungi can lessen the efficacy of copper-containing wood preservatives for wood products in-service. The purpose of this study was to evaluate wood mass loss and differential expression of three genes that have putative annotations for copper-transporting ATPase pumps (FIBRA_00974, FIBRA_04716, and FIBRA_01430). Untreated southern pine (SP) and SP treated with three concentrations of ammoniacal copper citrate (CC, 0.6, 1.2, and 2.4%) were exposed to two copper-tolerant Fibroporia radiculosa isolates (FP-90848-T and L-9414-SP) and copper-sensitive Gloeophyllum trabeum isolate (MAD 617) in a 4-week-long standard decay test (AWPA E10-19). Decay of copper-treated wood was inhibited by G. trabeum (p = 0.001); however, there was no inhibition of decay with increasing copper concentrations by both F. radiculosa isolates. Initially, G. trabeum and one F. radiculosa isolate (L-9414-SP) highly upregulated FIBRA_00974 and FIBRA_04716 on copper-treated wood at week 1 (p = 0.005), but subsequent expression was either not detected or was similar to expression on untreated wood (p = 0.471). The other F. radiculosa isolate (FP-90848-T) downregulated FIBRA_00974 (p = 0.301) and FIBRA_04716 (p = 0.004) on copper-treated wood. FIBRA_01430 expression by G. trabeum was not detected, but was upregulated by both F. radiculosa FP-90848-T (p = 0.481) and L-9414-SP (p = 0.392). Results from this study suggest that all three test fungi utilized different mechanisms when decaying copper-treated wood. Additionally, results from this study do not provide support for the involvement of these putative gene annotations for copper-transporting ATPase pumps in the mechanism of copper-tolerance.
Introduction
Copper is an essential metal for eukaryotic life (Lattore et al., 2011; Smith et al., 2017; Raffa et al., 2019). It functions in generating energy, mobilizing iron, inactivating enzymes, interacting and stabilizing ligands, and generating and detoxifying reactive oxygen species (ROS; Li et al., 2005; Smith et al., 2017; Raffa et al., 2019). Fungal cells uptake copper through ion exchange which permeates copper throughout and leads to accumulation inside the cell (Somers, 1963). Affinity, distribution, and accumulation of copper vary among fungal species. Fungi must strictly regulate copper levels as excess can cause oxidative damage, enzyme inhibition, interruptions in nutrient transport, protein and enzyme denaturation, or cellular death (Freeman and McIntyre, 2008; Smith et al., 2017).
Fungi utilize several mechanisms to deal with toxic metals in their immediate environment (Gadd, 1993; Hall, 2002). These metal tolerance mechanisms of fungi appear to include: efflux and excretion of accumulated metals (Weissman et al., 2000; Ito et al., 2007); cell wall complexation facilitated by extracellular polymeric slime materials which aid in controlling metal uptake (Daniel and Nilsson, 1989; Yahaya et al., 2009); chelation and/or precipitation by soluble metabolites (oxalic acid; Clausen, 2000; Clausen and Green, 2003; Green and Clausen, 2003; Jarosz-Wilkolazka and Graz, 2006; Tang et al., 2013); accumulated metal complexation and immobilization by metallothioneins (Schwartz et al., 2013); and metal sequestration to the periplasm or vacuoles (Pearce and Sherman, 1999). Each of these mechanisms can be employed singularly or in concert to prevent metal toxicity by reducing intracellular damage typically caused by oxidative species resulting from reactive metal ions.
Some decay fungi are able to successfully colonize and destroy copper-treated wood (DeGroot and Woodward, 1999; Hall, 2002; Green and Clausen, 2003). Copper-tolerant fungi thrive in the toxic environment of treated wood due to their ability to render copper ions found in treated wood inert (Gadd, 1993; Hastrup et al., 2005). Many copper-tolerant fungi produce high amounts of oxalic acid which precipitates copper ions as insoluble copper-oxalate and has been proposed to explain how these copper-tolerant fungal organisms overcome unfavorable copper environments (Shimanzono and Takubo, 1952; DeGroot and Woodward, 1999; Pohleven et al., 1999; Humar et al., 2002; Green and Clausen, 2003, 2005; Hastrup et al., 2005, 2006; Freeman and McIntyre, 2008; Arango et al., 2009; Schilling and Inda, 2011; Tang et al., 2013). It is hypothesized that high extracellular oxalic acid accumulation aids in the precipitation of copper ions into insoluble copper oxalate crystals, inactivating copper in this process (Gadd, 1993; DeGroot and Woodward, 1999; Green and Clausen, 2003; Hastrup et al., 2005; Freeman and McIntyre, 2008). Additionally, it has been documented that copper-tolerant fungi produce two to 17 times more oxalic acid when exposed to copper-treated wood compared to non-treated wood (Green and Clausen, 2005; Jenkins, 2012; Ohno et al., 2015).
Several studies including Postia placenta, Antrodia vaillantii, Fomitopsis (Tyromyces) palustris, Meruliporia (Poria) incrassata, Wolfoporia cocos, and F. radiculosa revealed the presence of copper-oxalate moolooite crystals produced on copper-treated wood (Murphy and Levy, 1983; Sutter et al., 1983, 1984; Tsunoda et al., 1997; DeGroot and Woodward, 1999; Clausen et al., 2000; Tang et al., 2013). Subsequent gene expression analysis showed that increased oxalate production in the presence of a copper-based preservative was regulated by increased expression of key genes in glyoxylate ametabolism (Tang et al., 2013). Hence, it has become generally accepted that oxalic acid is integral to the mechanism of copper-tolerance (Murphy and Levy, 1983; Sutter et al., 1983; Daniel, 1994; Leithoff et al., 1995; Pohleven et al., 2002; Green and Clausen, 2003, 2005; Hastrup et al., 2006; Freeman and McIntyre, 2008; Arango et al., 2009; Schilling and Inda, 2011; Tang et al., 2013). However, direct availability of oxalic acid, its accumulation, or copper oxalate precipitation does not always correlate with increased mass loss in copper-treated wood (Sutter et al., 1983; DeGroot and Woodward, 1999; Clausen, 2000; Schilling and Jellison, 2005; Ohno et al., 2017). Additionally, the effect of sodium oxalate added to non-treated and copper-treated wood does not increase mass loss caused by copper-tolerant fungi; therefore, it was suggested that oxalic acid accumulation and precipitation are not the exclusive method copper-tolerant fungi utilize in overcoming high copper concentrations (Schilling and Jellison, 2005; Ohno et al., 2017).
Copper-transporting ATPases are evolutionarily conserved proteins that mediate import, distribution, sequestration, and export of cellular copper (Migocka, 2015). In eukaryotes, ATPases are found in intracellular compartments, the Golgi membrane or the plasma membrane (depending on their primary function) serve in metal detoxification and shuttles copper to copper dependent enzymes in the secretory pathway (Gupta and Lutsenko, 2012). In yeast, copper homeostasis is not universal and specific proteins differ among genera. In Saccharomyces cerevisiae, high intracellular copper levels are sensed by transcription factor Ace1, which activates metallothioneins, Cup1 and Crs5, and a superoxide dismutase (Sod1; Raffa et al., 2019). In Cryptococcus neoformans, high intracellular copper levels are sensed by Cuf1, which activates metallothioneins, Cmt1 and Cmt2 to bind excess copper (Raffa et al., 2019). In Candida albicans, high intracellular copper levels are sensed by Cup2, which activates a copper exporter, Crp1, and metallothioneins Cup1 and Crd2 remove or bind excess copper (Raffa et al., 2019). In C. albicans, copper-transporting ATPase pumps are responsible for preventing intracellular copper concentrations from becoming toxic (Weissman et al., 2000).
In Fibroporia radiculosa (Peck) Glib & Ryvarden, three genes with function related to regulating copper concentrations were detected (Tang et al., 2012). Tang et al. (2012) determined that of the three putative gene annotations for ATPases, only one (FIBRA_00974) encoded a signal peptide and another (FIBRA_01430) had no homolog in copper-tolerant brown-rot decay fungus Serpula lacrymans, which could be indicative of the higher tolerance observed in F. radiculosa. Data for involvement of these genes in regulating copper tolerance, however, were conflicting (Tang et al., 2013). Although transcriptomics analysis suggested involvement of one COP-encoding gene (FIBRA_01430), the follow-up experiment using qRT-PCR showed this gene was not upregulated during decay of wood treated with a copper-based preservative compared to untreated wood (Tang et al., 2013). Jenkins (2012) measured expression of FIBRA_01430 in four isolates of F. radiculosa on copper-treated wood over an 8 week period and found expression was greater in the presence of copper. Based on these findings, it was hypothesized that this ATPase gene functions to prevent accumulation of toxic concentrations of copper in the cell during the early stages, and that this is a necessary step in initial colonization of copper-treated wood by F. radiculosa (Jenkins, 2012).
The goal of this study was to expand our current understanding of the genes thought to be involved in copper-tolerance of F. radiculosa during initial decay. We examined the differential expression of three putative copper transporting ATPase genes (FIBRA_00974, FIBRA_04716, and FIBRA_01430) in two copper-tolerant F. radiculosa isolates (FP-90848-T and L-9414-SP) and copper-sensitive Gloeophyllum trabeum (pers. Ex Fr.) Murr. (MAD 617) during decay of untreated and copper-treated SP wood. Implications from this study could specify genes responsible for overcoming unfavorable copper environments in copper-tolerant decay fungi.
Materials and Methods
Fungal Cultures
Two copper-tolerant F. radiculosa isolates (FP-98048-T and L-9414-SP; USDA-NRS-FMHC, Forest Products Laboratory, Madison, WI, United States) were used. One copper-sensitive G. trabeum (MAD 617; USDA-NRS-FMHC, Forest Products Laboratory, Madison, WI, United States) was also used. Fungal cultures were maintained on malt extract agar (BD, ThermoFisher Scientific, Waltham, MA, United States) at 27°C and 70% relative humidity (RH).
Preservative Treatment and Decay Tests
Experimental set-up consisted of accelerated decay tests following the American Wood Protection Association Standard E10-19 (American Wood Protection Association, 2019). SP wood wafers (40 by 30 by 3 mm) in soil block jars (one wafer/jar) were initially colonized for 3 weeks after inoculation by fungal mycelial plugs. SP test blocks (20 mm cubes) were vacuum-treated with three concentrations of ammoniacal copper citrate: 0.6% (3.350 kg Cu/m3), 1.2% (6.888 kg Cu/m3), and 2.4% (13.642 kg Cu/m3). Untreated SP blocks were included. After treatment, blocks dried under a hood overnight and were conditioned to 27°C and 30% RH for 2 weeks. Wood block exposure to the fungi was accomplished by aseptically placing two test blocks (steam sterilized 20 min, 122°C and cooled to room temperature) onto the colonized wood wafer in each glass bottle for each fungus and time point. Bottles were then incubated at 27°C and 70% RH. Weekly for each of 4 weeks, one test block from each of three glass bottles for each treatment was used to measure mass loss (n = 3). Test blocks were lightly brushed free of mycelia, oven-dried overnight (60°C), and reconditioned (27°C and 30% RH) for 2 weeks prior to weighing and calculating mass loss (%). The second test block from the same jar was used for RNA extractions (n = 3).
Sample Preparation and RNA Extraction
Approximately, 0.2 g of sawdust was produced by drilling into each test block with a sterile bit and placed into individual microtubes, and RNA was immediately extracted. The Ambion® RNAqueous™ Kit (ThermoFisher Scientific) was used to isolate RNA following the manufacturer’s specifications with an added DNaseI (Promega Corporation, Madison, WI, United States) digestion to removed unwanted genomic DNA contamination. DNaseI digestion was prepared according to the manufacturer’s specifications and was included between the Wash Solution 1 step and Wash Solution 2 step of the RNAqueous™ Kit. RNA for all samples was quantified by a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific). Yields ranged from 2 to 12 ng g−1 sawdust. RNA samples were stored at −80°C.
Protein Extraction and Western Blot
Fungal hyphae of the two F. radiculosa isolates were grown in 50 ml Bailey’s minimal media with 1% cellobiose and 2.5% glucose for 9 days at 27°C and 70% RH (Highley, 1973). Three replicate cultures were grown in untreated Baileys minimal media, Bailey’s minimal media with 3 mM copper sulfate, and Bailey’s minimal media with 1 mM sodium orthovanadate, an ATPase inhibitor. Whole protein extractions were conducted using a modified version of Zhang et al. (2011). Fungal hyphae (0.1 g) were collected by vacuum filtration and then homogenized in 1 ml of 2 M lithium acetate using an Omni probe homogenizer (3 × 5 s pulses; Omni International, Kennesaw, GA, United States). Homogenate was held at room temperature for 10 min and then centrifuged at 4,000 × g for 5 min at 4°C. The pellet was resuspended in 1 ml of 0.4 M sodium hydroxide by vortexing 5 s and then held on ice for 5 min. The homogenate was then centrifuged 10,000 × g for 5 min at 4°C. The pellet was resuspended in SDS buffer (1% SDS, 100 mM Tris-HCL pH 7, 1 mM protease inhibitor), by mixing with the pipet tip and then boiled 5 min at 100°C. Finally, this homogenate was centrifuged 4,000 × g for 5 min at room temperature and pellet was discarded. Protein fractions were quantified using the RC DC™ Protein Assay (Bio-Rad Laboratories) following the manufacturer’s specifications.
Protein fractions (0.5–5 μg) were loaded onto a NuPAGE™ 4–12% Bis-Tris polyacrylamide gel (ThermoFisher Scientific) and run in NuPAGE™ MOPS SDS buffer (ThermoFisher Scientific) at 100 V for 50 min. Resolved proteins were transferred to a nitrocellulose membrane (Bio-Rad Laboratories) in NuPAGE™ Transfer buffer (ThermoFisher Scientific) at 30 V for 60 min. The membrane was blocked with 3% Blotting Grade Blocker (Bio-Rad Laboratories) for 60 min and probed with ATP7A raised toward a human copper-translocating P-type ATPase protein (Aviva Systems Biology, San Diego, CA, United States) at a 1:500 dilution overnight. Secondary probing with Goat Anti-Rabbit IgG (H + L)-HRP conjugate (Bio-Rad Laboratories) at a 1:3000 dilution for 60 min followed. Blots were developed using the Immun-Blot® Opti-4CN Colorimetric Goat-anti-Rabbit kit (Bio-Rad Laboratories) following the manufacturer’s specifications and visualized on a Gel Doc XR+ system (Bio-Rad Laboratories). A positive control (EGY48 yeast lysate) for ATP7A was included (Santa Cruz Biotechnology, Inc., Dallas, TX, United States).
First Strand cDNA Synthesis
RNA (20 ng per 20 μl reaction mixture) was synthesized to first strand complementary DNA (cDNA) using the SuperScript™ II Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s specifications. cDNA synthesis was carried out on an MJ Research PTC-225 Thermal Cycler (Bio-Rad Laboratories, Carlsbad, CA, United States) with the following settings: incubation at 42°C for 50 min and inactivation at 70°C for 15 min. Both non-template controls and reverse transcriptase free controls were included. cDNA samples were stored at −20°C.
qPCR Analysis
Gene-specific primers were designed using gene and coding sequences for FIBRA_00974, FIBRA_04716, and FIBRA_01430 in F. radiculosa through NCBI GenBank. For the transcripts, primer pairs spanned at least one intron. Amplicon lengths ranged from 299 to 900 bp depending on the gene (FIBRA_00974: 649 bp; FIBRA_04716: 299 bp; and FIBRA_01430: 900 bp). Table 1 lists the primer sequences for the target genes and endogenous control. A preliminary electrophoresis analysis was performed from RT-PCR products for each gene and non-template and reverse-transcriptase-free controls on a 2% agarose gel in TAE (1x) buffer. Target bands were verified based on amplicon length for the respective genes. Additionally, target bands were absent from controls but present in samples for the respective amplicon lengths.

Table 1. Primer sequences for FIBRA_00974, FIBRA_04716, and FIBRA_01430 genes and β-actin (housekeeping gene) designed for this study.
Expression profiles of FIBRA_00974, FIBRA_04716, and FIBRA_01430 were monitored in real time using a MyiQ™ 5 Real-Time Detection System (Bio-Rad Laboratories). Sample preparations (undiluted cDNA) were carried out using the iQ™ SYBR® Green Supermix (Bio-Rad Laboratories) following the manufacturer’s specifications. Three biological replicates were run in duplicate for each plate against the non-template and reverse transcriptase free controls for each of the treatments, time points, and genes. The reaction protocol included an initial denaturation at 95°C for 3 min, followed by 40 cycles of a 15 s 95°C denaturation, 30 s annealing at 60°C, and a 30 s 72°C extension. For FIBRA_01430, annealing was carried out at 55°C. A melt curve analysis of the resulting PCR product was performed immediately following amplification to check for primer specificity and primer dimer formation (1 min incubation at 95°C, followed by a 1 min annealing at 55°C prior to 81 cycles of 10 s with temperatures increasing by 0.5°C each cycle).
Using the housekeeping gene (β-actin; 270 bp amplicon) mean CT values, average replicate CT values were normalized for all plates. Normalized CT values were used to calculate mean fold change (2-ΔΔCT) in expression of the three ATPase genes using the comparative CT method (Livak and Schmittgen, 2001). For the three test fungi, untreated wood at week 1 was used as the endogenous control for fold change calculations for FIBRA_00974, FIBRA_04716, and FIBRA_01430. Mean fold changes were then converted to the log2 scale to equally represent upregulation (values > 0) and downregulation (values < 0). Values reflect the differences between the endogenous control and each treatment in the subsequent weeks. If no value is shown in the subsequent figures, the gene was not expressed for that particular test variable.
Statistical Analysis
A three-way ANOVA was performed using the general linear model (GLM) to determine effects of fungi, treatment, and time on mass loss. GLM describes statistical relationships between multiple factors using mean values to find significant differences. Mass loss data were reported in percentages, therefore, did not meet the assumptions of a parametric statistical analysis. Data were analyzed after applying the arcsine of the square root transformations to proportions.
Non-parametric statistical analysis using the Kruskal-Wallis test (p = 0.05) was conducted to determine differences among treatment and time for FIBRA_00974, FIBRA_04716, and FIBRA_01430 expression, respectively (Goni et al., 2009). Treatments were grouped for each fungus prior to statistical analysis. Additionally, time points were grouped for each fungus prior to statistical analysis. Statistical analyses were performed using Minitab 17.2.1 (Minitab, LLC, State College, PA, United States).
Results
Decay
Mass loss data (%) differed among the three test fungi and on untreated and treated wood over time (Figure 1). ANOVA showed significant effects of fungus, treatment, time, and all interactions (values of p ≤ 0.001). Gloeophyllum trabeum indicated copper sensitivity on copper-treated wood through 4 weeks (Figure 1A). On untreated wood, G. trabeum increased mass loss from 1.2% at week 1 to 11.1% at week 4. On copper-treated wood, G. trabeum produced significantly lower mass loss for all copper levels, < 2.4% (p = 0.001). In contrast, F. radiculosa FP-90848-T (Figure 1B) and L-9414-SP (Figure 1C) were not inhibited by copper-treated wood through 4 weeks. Mass losses caused by both F. radiculosa isolates on untreated wood were significantly lower (< 9%) than mass losses of copper-treated wood (p = 0.000). Fibroporia radiculosa FP-90848-T caused significantly greater mass losses on all concentrations of copper-treated wood compared to untreated wood by week 4, up to 12.6% (p = 0.001). Fibroporia radiculosa L-9414-SP caused significantly greater mass losses on all concentrations of copper-treated wood compared to untreated wood by week 4, up to 16.4% (p = 0.001). However, decay by F. radiculosa L-9414-SP was less on 2.4% CC by week 4 compared to FP-90848-T.
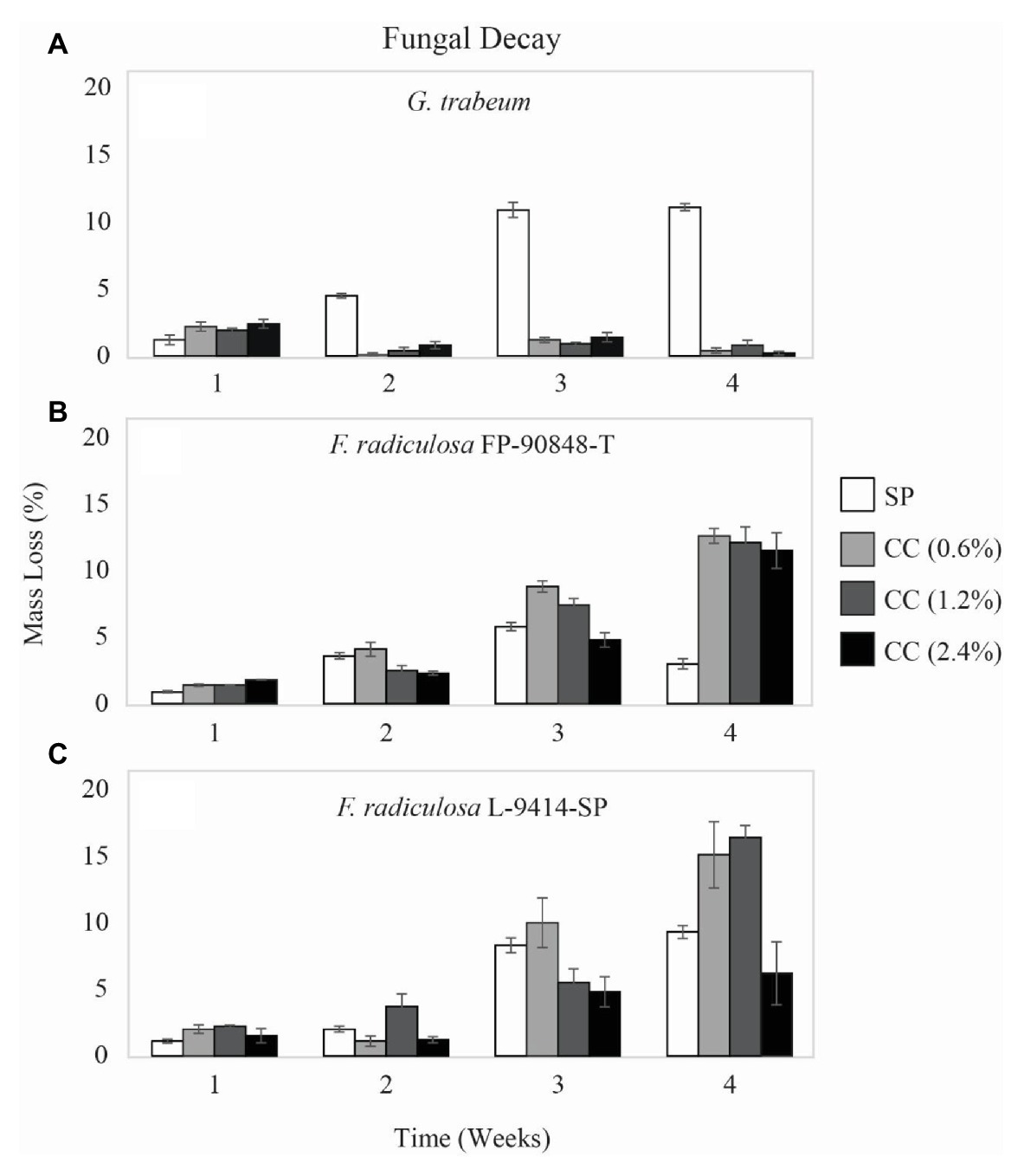
Figure 1. Mass loss (%) ± SE of untreated (SP), 0.6% ammoniacal copper citrate treated (0.6% CC), 1.2% ammoniacal copper citrate treated (1.2% CC), and 2.4% ammoniacal copper citrate treated (2.4% CC) southern pine (SP) test blocks exposed to: (A) copper-sensitive Gloeophyllum trabeum (MAD 617), (B) copper-tolerant Fibroporia radiculosa (FP-90848-T), and (C) copper-tolerant Fibroporia radiculosa (L-9414-SP) over the course of 4 weeks (n = 3).
Western Blot
Whole cell lysate protein fractions probed with an antibody raised to human ATP7A can be seen in Figure 2. Results show reactivity in both F. radiculosa isolates at 55 and 40 kDa when grown in Bailey’s minimal media and Bailey’s minimal media with 3 mM copper sulfate. Three bands were seen in both F. radiculosa isolates. Two bands at 55 kDa and a third band around 40 kDa. The three bands could correspond to the three putative copper-transporting ATPases characterized in F. radiculosa. There was no ATP7A reactivity when grown in Bailey’s minimal media with 1 mM of sodium orthovanadate, an inhibitor of ATPases, for both F. radiculosa isolates.
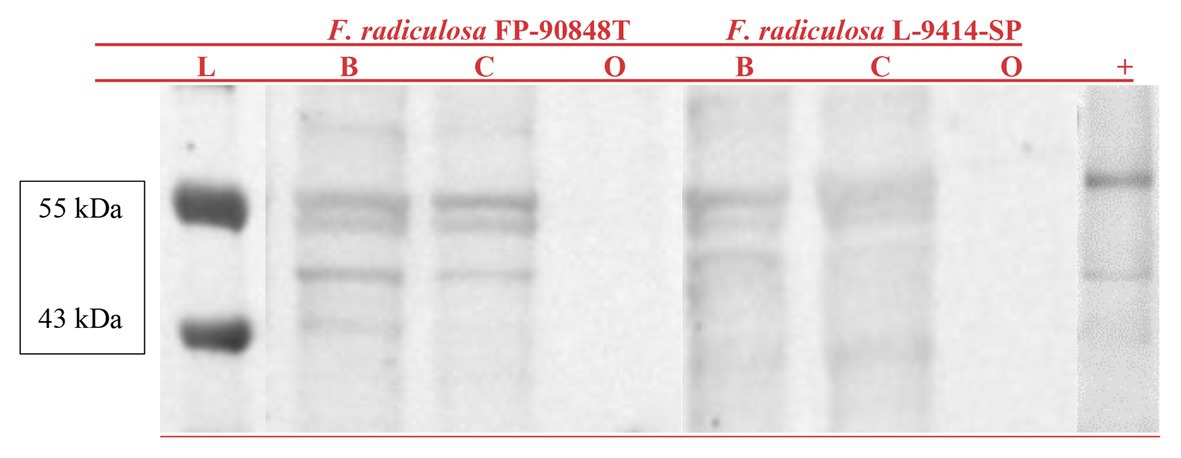
Figure 2. Western blot of ATP7A probed whole cell lysate protein fractions from Fibroporia radiculosa isolates FP-90848-T and L-9414-SP grown in Bailey’s minimal media (B) Bailey’s minimal media containing 3 mM copper sulfate (C), and Bailey’s minimal media with 1 mM sodium orthovanadate (O). A molecular marker (L) is represented on the left. An EGY48 yeast lysate with ATP7A reactivity is represented on the right (+).
FIBRA_00974
Expression of a copper-transporting ATPase gene (FIBRA_00974) differed among the three test fungi exposed to untreated and copper-treated wood (Figure 3). Gloeophyllum trabeum (Figure 3A) upregulated FIBRA_00974 significantly more when exposed to copper-treated wood compared to untreated wood (p = 0.000). Additionally, there was no statistically significant difference of G. trabeum FIBRA_00974 expression for weeks 1–3 (p = 0.944). There was no detectable FIBRA_00974 expression for G. trabeum on untreated and copper-treated wood at week 4 despite housekeeping gene amplification.
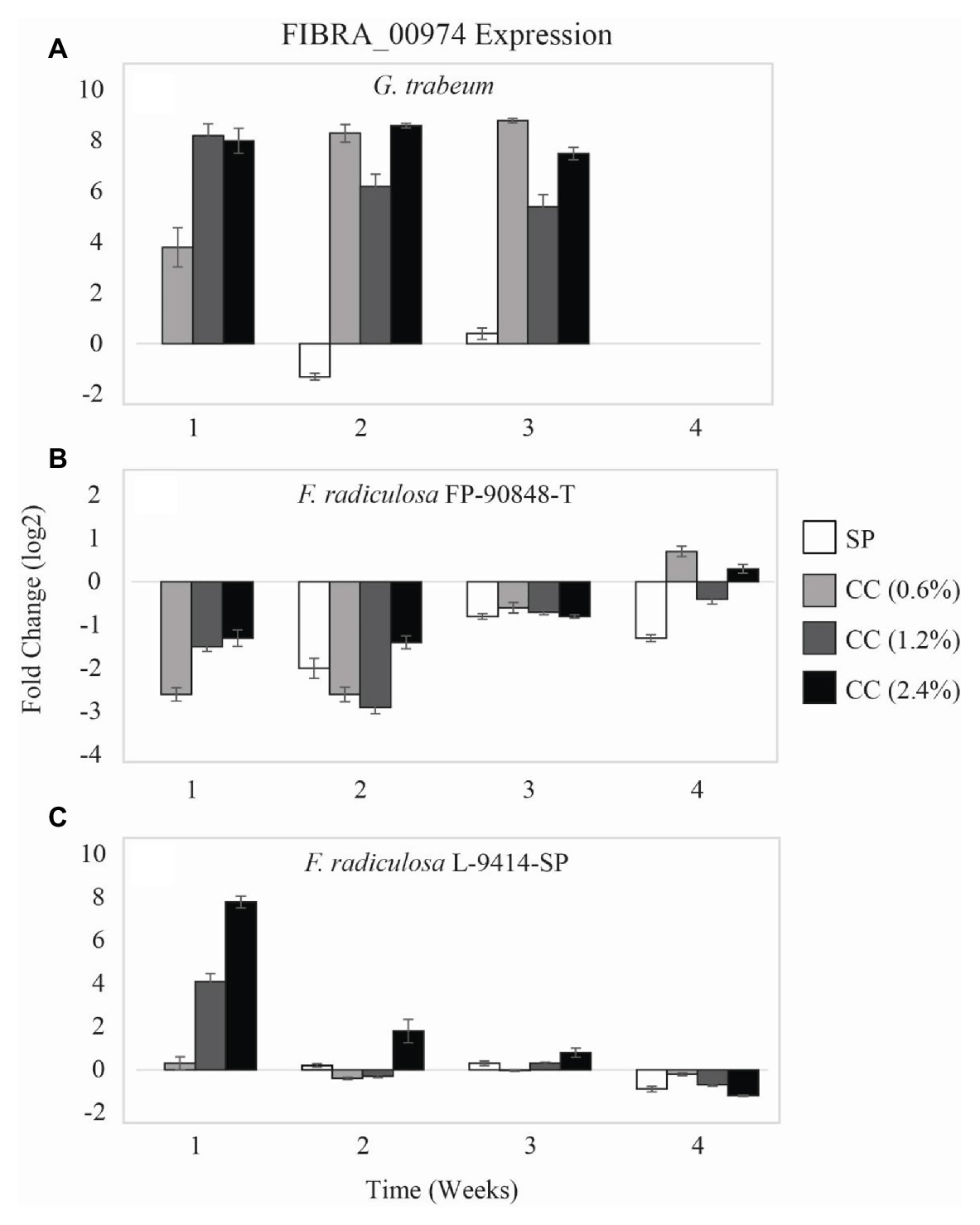
Figure 3. Gene expression values (log2) ± SE of the copper-transporting FIBRA_00974 gene of: (A) copper-sensitive G. trabeum (MAD 617), (B) copper-tolerant F. radiculosa (FP-90848-T), and (C) copper-tolerant F. radiculosa (L-9414-SP) on untreated (SP), 0.6% ammoniacal copper citrate treated (0.6% CC), 1.2% ammoniacal copper citrate treated (1.2% CC), and 2.4% ammoniacal copper citrate treated (2.4% CC) SP test blocks over the course of 4 weeks. Values above 0 indicate upregulation. Values below 0 indicate downregulation relative to expression on SP at week 1 for each fungus, respectively (set at 0). FIBRA_00974 was not expressed for G. trabeum at week 4.
The two F. radiculosa isolates differed in FIBRA_00974 expression over the course of this study. Generally, F. radiculosa FP-90848-T downregulated FIBRA_00974 on untreated and copper-treated wood (Figure 3B). However, there was minimal FIBRA_00974 upregulation when F. radiculosa FP-90848-T was exposed to 0.6 and 2.4% CC at week 4 compared to the other weeks. Additionally, there was no statistically significant difference of F. radiculosa FP-90848-T FIBRA_00974 expression between untreated and copper-treated wood (p = 0.301). Fibroporia radiculosa FP-90848-T downregulated FIBRA_00974 more at week 2 compared to the other weeks (p = 0.000). Fibroporia radiculosa L-9414-SP upregulated FIBRA_00974 more on copper-treated wood compared to untreated wood at week 1 (Figure 3C). Additionally, FIBRA_00974 upregulation increased with increasing copper concentration at week 1. However, the grouped analysis showed no differences in FIBRA_00974 expression for this isolate on untreated and copper-treated wood (p = 0.471). Fibroporia radiculosa L-9414-SP upregulated FIBRA_00974 more at week 1 compared to the other weeks and downregulated FIBRA_00974 more at week 4 compared to the other weeks (p = 0.000).
FIBRA_04716
A second copper-transporting ATPase gene (FIBRA_04716) differed in expression among the three test fungi on untreated and copper-treated wood (Figure 4). Gloeophyllum trabeum (Figure 4A) upregulated FIBRA_04716 more on copper-treated wood compared to untreated wood at week 1 (p = 0.005). After week 1, G. trabeum had no detectable FIBRA_04716 expression on untreated and copper-treated wood.
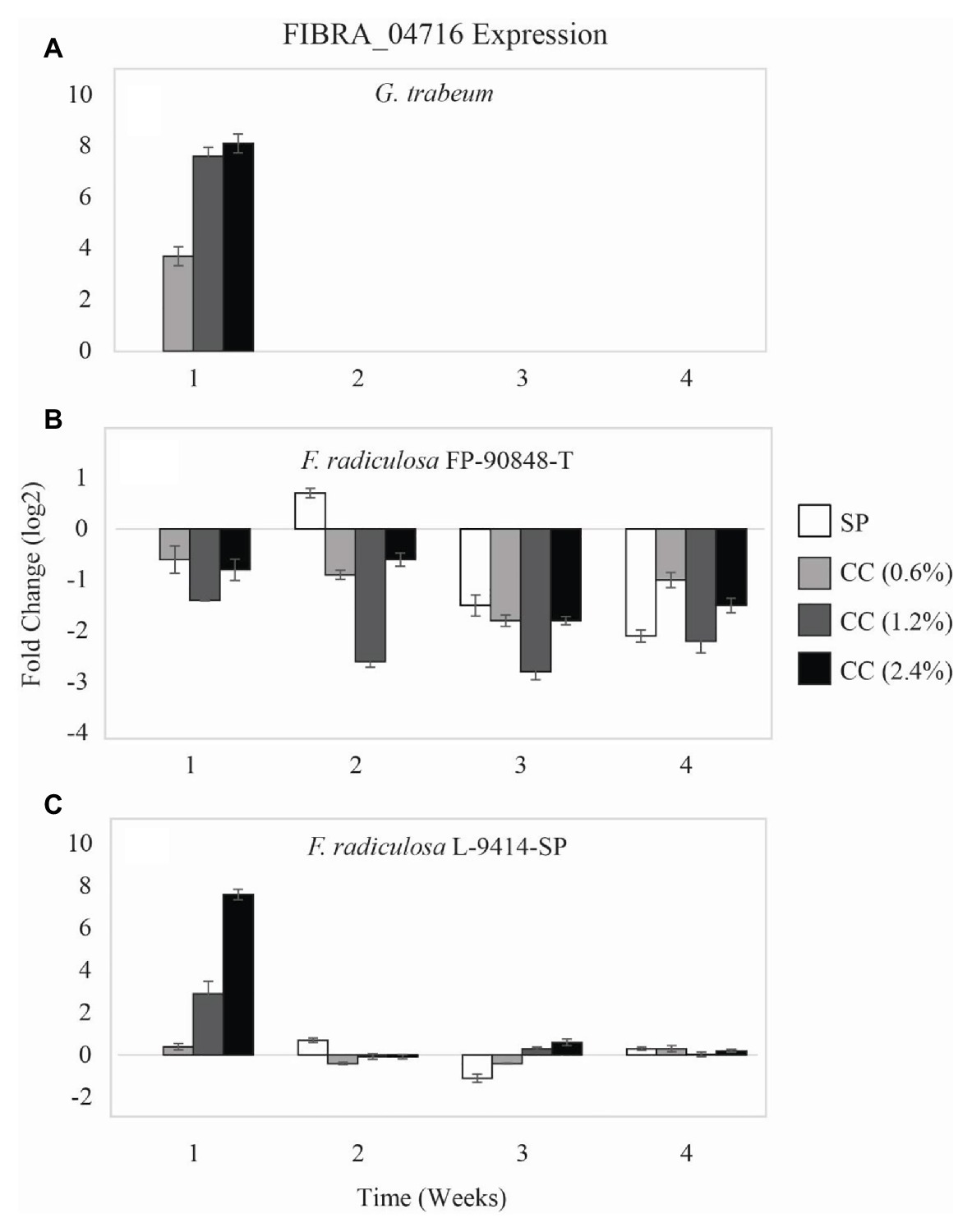
Figure 4. Gene expression values (log2) ± SE of the copper-transporting FIBRA_04716 gene of: (A) copper-sensitive G. trabeum (MAD 617), (B) copper-tolerant F. radiculosa (FP-90848-T), and (C) copper-tolerant F. radiculosa (L-9414-SP) on untreated (SP), 0.6% ammoniacal copper citrate treated (0.6% CC), 1.2% ammoniacal copper citrate treated (1.2% CC), and 2.4% ammoniacal copper citrate treated (2.4% CC) SP test blocks over the course of 4 weeks. Values above 0 indicate upregulation. Values below 0 indicate downregulation relative to expression on SP at week 1 for each fungus, respectively (set at 0). FIBRA_04716 was not expressed for G. trabeum at weeks 2–4.
Again, the FIBRA_04716 expression patterns of the two F. radiculosa isolates differed over the course of this study. Fibroporia radiculosa FP-90848-T downregulated FIBRA_04716 (Figure 4B). This isolate had greater FIBRA_04716 downregulation on copper-treated wood compared to untreated wood at week 1 through week 3 (p = 0.004). However, by week 4, F. radiculosa FP-90848-T downregulated FIBRA_04716 more on untreated wood compared to copper-treated wood. Additionally, this isolate downregulated FIBRA_04716 more at week 3 compared to the other weeks (p = 0.001). Generally, F. radiculosa L-9414-SP showed FIBRA_04716 upregulation on untreated and copper-treated wood over the 4 weeks (Figure 4C); however, there were instances where FIBRA_04716 was downregulated (copper-treated wood, week 2). Also it should be mentioned at week 1, F. radiculosa L-9414-SP upregulated FIBRA_04716 more on copper-treated wood and increased upregulation with increasing copper concentration. These results are comparable to FIBRA_00974 expression at week 1 seen in this isolate, and to FIBRA_00974 and FIBRA_04716 expression results of G. trabeum. Despite having greater FIBRA_04716 upregulation at week 1 on copper-treated wood, the grouped analysis for this isolate on untreated and copper-treated wood lacked statistical significance in FIBRA_04716 expression (p = 0.059). Fibroporia radiculosa L-9414-SP upregulated FIBRA_04716 more at week 1 compared to the other weeks (p = 0.048).
FIBRA_01430
Two of the three test fungi differed in expression of a third copper-transporting ATPase gene (FIBRA_01430) on untreated and copper-treated wood (Figure 5). Gloeophyllum trabeum had no detectable expression of FIBRA_01430 on untreated and copper-treated wood over the course of this study thus no data are shown. Unlike FIBRA_00974 and FIBRA_04716 expression patterns, both F. radiculosa isolates (FP-90848-T and L-9414-SP) upregulated FIBRA_01430 on untreated and copper-treated wood (p = 0.481 and p = 0.392, respectively). However, F. radiculosa FP-90848-T (Figure 5A) increased in FIBRA_01430 upregulation on 0.6 and 2.4% copper-treated wood over time (p = 0.000).
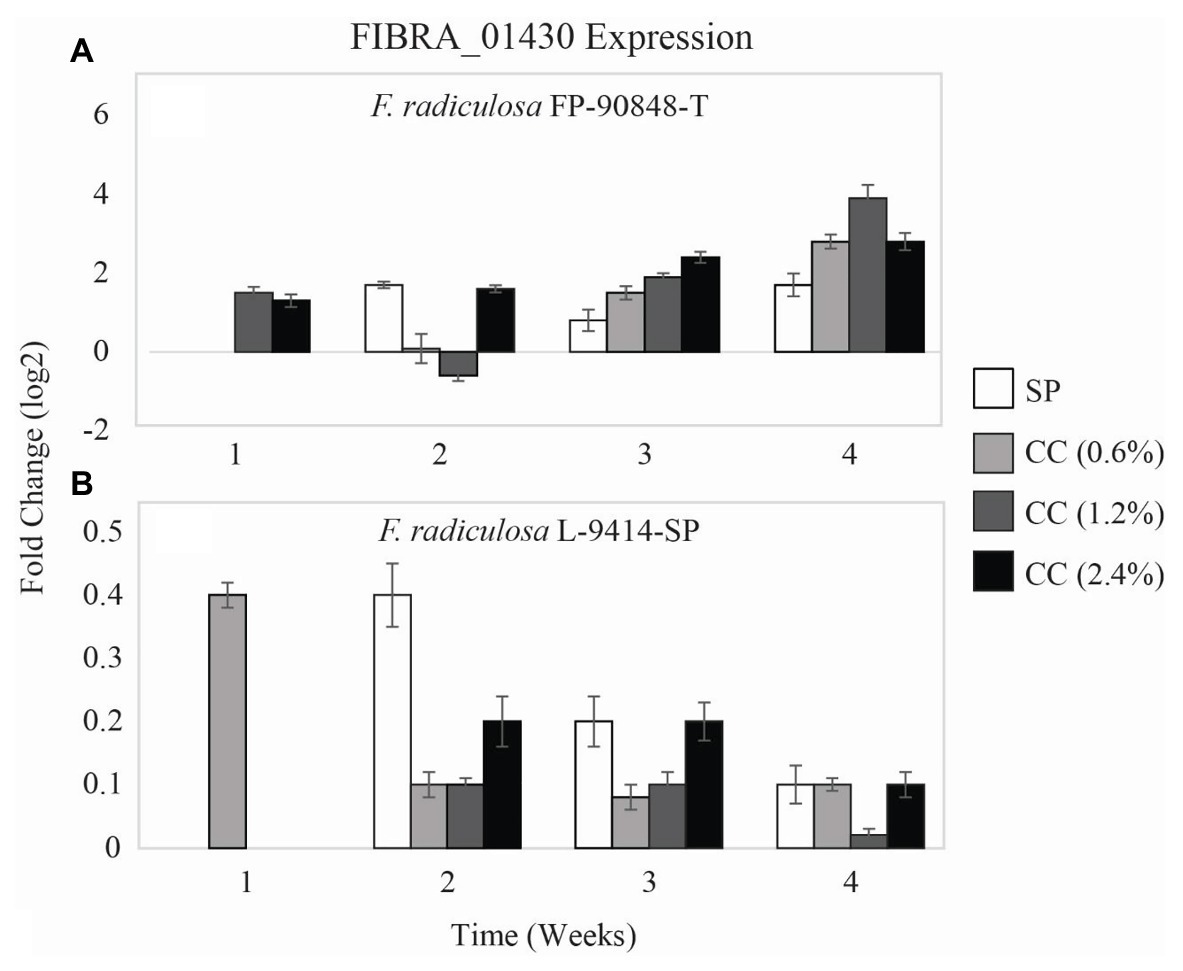
Figure 5. Gene expression values (log2) ± SE of the copper-transporting FIBRA_01430 gene of: (A) copper-tolerant F. radiculosa (FP-90848-T) and (B) copper-tolerant F. radiculosa (L-9414-SP) on untreated (SP), 0.6% ammoniacal copper citrate treated (0.6% CC), 1.2% ammoniacal copper citrate treated (1.2% CC), and 2.4% ammoniacal copper citrate treated (2.4% CC) SP test blocks over the course of 4 weeks. Values above 0 indicate upregulation. Values below 0 indicate downregulation relative to expression on SP at week 1 for each fungus, respectively (set at 0). FIBRA_01430 was not expressed for G. trabeum at weeks 1–4, F. radiculosa FP-90848-T on 0.6% CC at week 1, and F. radiculosa on 1.2 and 2.4% CC at week 1.
Discussion
To extend the service life of treated-wood products, it is critical to determine the mechanisms by which decay fungi can deteriorate wood. Due to the Environmental Protection Agency’s restriction of chromated copper arsenate (CCA) in 2004, arsenic-free and chromium-free preservatives utilize copper as the primary means of protection against biological organisms (Lebow, 2004, 2010; Lebow et al., 2004; Freeman and McIntyre, 2008). Copper-tolerant brown-rot fungi are quite problematic because of their ability to degrade copper-treated wood products in service. Copper tolerance necessitates the typical inclusion of co-biocides in commercial preservatives treatments (Illman et al., 2000). But to better understand how F. radiculosa responds to copper without co-biocides, we used the model copper preservative, copper citrate (Clausen et al., 2000; Clausen and Green, 2003; Hastrup et al., 2005), for this study. In addition, we were interested in how F. radiculosa initiates decay; therefore, we chose to determine mass loss during early decay (weeks 1 through 4).
Mass loss results of this study are consistent with previous studies which determined that G. trabeum is significantly inhibited by copper-treated wood (Green and Clausen, 2003; Hastrup et al., 2005, 2006; Jenkins et al., 2014; Ohno et al., 2017). Humar et al. (2002) treated Norway spruce (Picea abies) with copper sulfate and copper octanoate with ethanolamine (CuE) and showed G. trabeum inhibition on both copper-treatments by 4 weeks (3.9 and 0.0% mass loss, respectively). Green and Clausen (2003) reported 0% mass loss of 1.2% CC-treated wood by G. trabeum at 10 weeks. Humar et al. (2006) found G. trabeum inhibition on Norway spruce treated with copper sulfate with potassium dichromate (CuCr) and copper sulfate with CuE at weeks 1, 2, and 4 with only 0.3% mass loss on CuCr and 0.4% mass loss on CuE by week 4. Hastrup et al. (2006) treated SP with 1.2% CC and showed 4.8% mass loss by G. trabeum at week 10. Liew and Schilling (2012) reported similar effects of copper CuE-treated and micronized copper quaternary-treated wood on G. trabeum by week 12 (4.9 and 2.1% mass loss, respectively). Jenkins et al. (2014) also found copper sensitivity of G. trabeum on 1.2% CC-treated wood at weeks 2 and 4; however, there was no copper sensitivity when SP was not pressure-treated with copper (i.e., dipped in copper, adjacent to copper). In each of these studies, the copper effects on decay by G. trabeum were similar to those of the present study.
Mass loss results of this study are also consistent with previous studies of decay of copper-treated wood by F. radiculosa. Clausen and Green (2003) reported similar mass loss by F. radiculosa (formerly Meruliporia incrassata) TFFH 294 on CC-treated wood for weeks 1 through 4 (1.6, 3.5, 3.6, and 8%, respectively). Results of Green and Clausen (2003) also showed high mass loss by F. radiculosa (formerly Antrodia radiculosa) FP-90848-T and F. radiculosa TFFH 294 on 1.2% CC-treated wood at 10 weeks (49 and 45%, respectively). They also reported mass loss to be greater on copper-treated wood compared to untreated wood for FP-90848-T and TFFH 294 (26 and 37%, respectively) at 10 weeks (Green and Clausen, 2003).
Among F. radiculosa isolates, there is diversity in timing and levels of gene expression. Differences in F. radiculosa isolates seen in this study were consistent with previous work suggesting F. radiculosa L-9414-SP has a delayed response in adapting to copper when compared to other F. radiculosa isolates (Jenkins, 2012; Ohno et al., 2015). Additional isolates of F. radiculosa (TFFH 294 and L-11659-SP) have been documented to have differences in oxalate accumulation and differential expression of genes encoding a citrate synthase, an isocitrate lyase, a glyoxylate dehydrogenase, a succinate/fumarate antiporter, and a copper transporting ATPase pump over the course of 8 weeks (Ohno et al., 2015). Zelinka et al. (2018) used synchrotron-based X-ray fluorescence microscopy to observe copper distribution in SP wood wafers that were dipped in copper sulfate pentahydrate after exposure to three copper-tolerant fungi, F. radiculosa, P. placenta, and F. (Tyromyces) palustris. After exposure to F. radiculosa for 9 weeks, there were much lower copper concentrations remaining in the dipped wafers, but only slightly lower copper concentrations after P. placenta and F. palustris exposure. They suggested that copper-tolerant species could rely on unique undefined processes for copper-tolerance. It has also been documented that brown-rot fungi in general have variations in the biochemical processes of decay and are highly dependent upon the environment in which they are grown (Presley and Schilling, 2017). From these studies mentioned above, it could be inferred that the necessary components in copper-tolerance are unique to a particular species and even isolate.
A possible mechanism of copper-tolerance is translocation of copper out of the area of fungal growth. Previous authors have reported that preservative tolerant fungi appear to be able to mobilize preservative elements, including copper, and decrease their concentration in the affected zone (Choi et al., 2012; Hastrup et al., 2014; Zelinka et al., 2018). Relatively few studies have investigated copper-tolerant F. radiculosa for other intracellular methods to overcome the toxic effects of copper (Tang et al., 2013; Ohno et al., 2015). Tang et al. (2013) reported minimal upregulation of one ATPase gene (FIBRA_01430) at week 3 when F. radiculosa was grown on wood treated with micronized copper quaternary. In addition, Tang et al. (2013) found no correlation between ATPase expression and mass loss. Ohno et al. (2015) reported upregulation of one ATPase gene (FIBRA_01430) at weeks 2 and 4 when F. radiculosa was grown on copper citrate-treated wood. From that study, Ohno et al. (2015) hypothesized that the mechanism of copper-tolerance is dependent on the intracellular management of copper facilitated by the copper-transporting ATPase genes in combination with extracellular oxalic acid accumulation and precipitation in F. radiculosa.
ATPase gene expression in other filamentous fungi is limited. Rhizophagus irregularis, an arbuscular mycorrhizal fungus, can tolerate high concentrations of metals in soils as a result of antioxidant defense activation. Tamayo et al. (2014) analyzed four ATPase encoded genes in R. irregularis, three characterized ATPase genes comparable to S. cerevisiae ATPases (i.e., shuttles copper ions to copper dependent enzymes) and one characterized as a plasma membrane ATPase comparable to C. albicans (i.e., copper detoxification). They showed upregulation of the three S. cerevisiae-like ATPases indicating a role in supplying copper to enzymes required for fungal establishment; however, they showed no expression data on the one C. albicans-like ATPase. In the corn pathogen, Cochliobus heterostrophus, Saitoh et al. (2009) analyzed heavy metal ATPase genes, which were divided into three groups. Group A, which delivers copper to copper-dependent proteins, and groups B and C, which are fungal specific and function in cell membrane copper efflux (Saitoh et al., 2009). However, this was just a phylogenetic analysis and no expression data were shown for the three groups characterized. Uldschmid et al. (2003) identified an ATPase protein, ctaA, in the white-rot fungus, Trametes versicolor. They showed ctaA was upregulated in the presence of copper and downregulated in the absence of copper and concluded this ATPase is localized to the Golgi membrane and functions in providing copper to copper-dependent enzymes, like laccase (Uldschmid et al., 2003).
The current study focused on analyzing intracellular ATPase genes to determine if they are necessary in the mechanism of copper-tolerance in F. radiculsoa. Copper-sensitive G. trabeum had greater FIBRA_00974 upregulation when grown on all copper-treatments (Figure 3A). In addition, G. trabeum also exhibited greater FIBRA_04716 upregulation when grown on all copper-treatments at week 1 (Figure 4A). These results suggest G. trabeum was attempting to manage the unfavorable copper environment, but ultimately was not successful, indicated by the lack of mass loss in copper-treated wood (Figure 1A).
The F. radiculosa isolates downregulated FIBRA_00974 (Figures 3B,C) and FIBRA_04716 (Figures 4B,C) after week 1, but upregulated FIBRA_01430 (Figures 5A,B). Additionally, both isolates showed no significant upregulation in expression of FIBRA_00974, FIBRA0416, and FIBRA_01430 after week 1 on copper-treated wood. These results suggest F. radiculosa does not rely upon intracellular copper management when exposed to increasing concentrations of copper. F. radiculosa still managed to cause decay on copper-treated wood (Figures 1B,C); however, this is likely not facilitated by the copper-transporting ATPase gene in F. radiculosa.
Gloeophyllum trabeum and F. radiculosa L-9414-SP had similar FIBRA_00974 (Figures 3A,C) and FIBRA_04716 (Figures 4A,C) upregulation on copper-treated wood at week 1. Gloeophyllum trabeum continued this trend in FIBRA_00974 upregulation at weeks 2 and 3 on copper-treated wood. Fibroporia radiculosa L-9414-SP also increased FIBRA_00974 and FIBRA_04716 upregulation with increasing copper-concentration. However, by week 2, there were no significant differences in FIBRA-00974 and FIBRA_0416 expression on untreated and copper-treated wood by this F. radiculosa isolate. This result is interesting and could suggest F. radiculosa L-9414-SP, like G. trabeum, responded similarly to an unfavorable copper environment at week 1. These results might suggest that FIBRA_00974 (and possibly FIBRA_04716) are involved in a mechanism of copper susceptibility in decay fungi; however, additional research is needed to address this hypothesis.
Expression of three copper-transporting ATPase genes thought to be involved in the mechanism of copper tolerance was quantified for the first time. Additionally, ATPase expression between copper-sensitive and copper-tolerant decay fungi during early decay of untreated and copper-treated wood had not been compared previously. This was one of a few studies in which intracellular methods for overcoming toxic copper concentrations in decay fungi were evaluated. Gloeophyllum trabeum had greater FIBRA_00974 and FIBRA_04716 upregulation when grown on copper-treated wood compared to untreated wood; however, this fungus was not successful at decay of copper-treated wood. Fibroporia radiculosa had similar ATPase expression on untreated and copper-treated wood (with the exception of FIBRA_00974 and FIBRA_04716 at week 1), and thus showed the same capacity to decay copper-treated wood as untreated wood (Figures 1B,C).
Environmental stresses such as metal toxicity (i.e., copper toxicity) enhance ROS production and threaten cell viability by causing a variety of issues (e.g., lipid peroxidation, protein oxidation, nucleic acid damage, enzyme inhibition, and programmed cell death; Sharma et al., 2012). To avoid oxidative injury, cells control ROS levels by means of non-enzymatic and enzymatic antioxidants. Enzymatic antioxidants include a wide variety of enzymes including superoxide dismutases, catalases, and peroxidases and increased activity of these enzymes deal with environmentally induced oxidative stress (Noctor and Foyer, 1998; Sharma et al., 2012). Increased metal tolerance has been linked to antioxidant capacity to scavenge toxic ROS and simultaneous expression of multiple antioxidant enzymes is more effective (Lee et al., 2007; Zaefyzadeh et al., 2009; Chen et al., 2010). In fact, it has been suggested that specific cellular responses to unfavorable environments are induced in decay fungi. These responses are: (a) production of laccases, peroxidases, and other lignin modifying enzymes (Peralta et al., 2017); (b) tolerance of oxidative stress and high levels of ROS facilitated by high levels of antioxidant enzymes like superoxide dismutase, catalase, and glutathione peroxidase an glutathione reductase (Belinky et al., 2003; Jaszek et al., 2006; Maciel et al., 2013); and (c) production of intracellular cytochrome p450 monooxygenases (Magan et al., 2010; Coelho-Moreira et al., 2013, 2018). It is highly possible that one or even several of these antioxidant enzymes could be implicated as critical components in the mechanism of copper-tolerance in decay fungi; however, additional research is needed to verify this statement. The mechanism of copper-tolerance in decay fungi remains unclear. Oxalic acid has been implicated in extracellular management of copper by decay fungi but intracellular management of copper by decay fungi remains unknown. Results of this research suggest copper-tolerant fungi like F. radiculosa utilizes a different mechanism to decay copper-treated wood when compared to copper-sensitive fungi like G. trabeum. Roles of the three copper-transporting ATPases in overcoming toxic copper environments in copper-tolerant fungi could not be confirmed, but differences were observed in gene expression between fungi and between untreated and copper-treated wood. Although this study did not pinpoint critical components of intracellular copper management by copper-tolerant brown-rot decay fungi, future studies can use the information provided to determine a detailed explanation of the mechanism of copper tolerance. Understanding this mechanism will prove critical in reducing the destruction of treated-wood products by copper-tolerant brown-rot fungi. Ultimately, additional research is needed to determine how F. radiculosa manages copper intracellularly. Additional studies should perhaps focus on analyzing gene expression without the wood substrate (i.e., liquid media) to better understand how certain interactions are happening. In addition, there are a multitude of stress-response genes (i.e., laccases, catalases, superoxide dismutases, cytochrome p450 monooxygenases, thioredoxins, etc.) that could play a role in copper-tolerance in wood decay fungi and should be investigated. Future research to comprehensively explain decay initiation by copper-tolerant brown-rot decay fungi could eventually lead to knowledge of how copper-tolerant brown-rot decay fungi manage toxic copper environments.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
AB contributed technical and laboratory assistance, and aided in the materials and methods write up. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
American Wood Protection Association (2019). Standard methods of testing wood preservatives by laboratory soil-block cultures. Standard E10-18. Book of Standards. American Wood-Preservers Association, Birmingham, AL, pp. 418–428.
Arango, R. A., Lebow, P., and Green, F. (2009). Correlation between oxalic acid production and tolerance of Tyromyces palustris strain TYP-6137 to N’,N-naphthaloylhydroxamine. Int. Biodeterior. Biodegrad. 63, 46–51. doi: 10.1016/j.ibiod.2008.05.006
Belinky, P. A., Flikhtein, N., Lechenko, S., Gepstein, S., and Dosoretz, C. G. (2003). Reactive oxygen species and induction of lignin peroxidase in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 69, 6500–6506. doi: 10.1128/AEM.69.11.6500-6506.2003
Chen, Q., Zhang, M., and Shen, S. (2010). Effect of salt on malondialdehyde and antioxidant enzymes in seedling roots of Jerusalem artichoke (Helianthus tuberosus L.). Acta Physiol. Plant. 62, 2599–2613. doi: 10.1007/s11738-010-0543-5
Choi, Y. S., Kim, J. J., Kim, M. J., Imamura, Y., Yoshimura, T., and Kim, G. H. (2012). Fungal biodegradation of CCA-treated wood and removal of its metal components. Chemosphere 88, 725–729. doi: 10.1016/j.chemosphere.2012.03.062
Clausen, C. A. (2000). CCA removal from treated wood using a dual remediation process. Waste Manag. Res. 18, 485–488. doi: 10.1177/0734242X0001800510
Clausen, C. A., and Green, F. (2003). Oxalic acid overproduction by copper-tolerant brown-rot basidiomycetes on southern yellow pine treated with copper-based preservatives. Int. Biodeterior. Biodegrad. 51, 139–144. doi: 10.1016/S0964-8305(02)00098-7
Clausen, C. A., Green, F., Woodward, B. M., Evans, J. W., and DeGroot, R. C. (2000). Correlation between oxalic acid production and copper tolerance in Wolfiporia cocos. Int. Biodeterior. Biodegrad. 46, 69–76. doi: 10.1016/S0964-8305(00)00044-5
Coelho-Moreira, J. S., Bracht, A., Souza, A. C. S., Oliveira, R. F., Sa-Nakanishi, A. B., Souza, C. G. M., et al. (2013). Degradation of diuron by Phanerochaete chrysosporium: role of ligninolytic enzymes and cytochrome P450. Biomed. Res. Int. 2013:251354. doi: 10.1155/2013/251354
Coelho-Moreira, J., Brugnari, T., Sa-Nakanishi, A., Castoldi, R., de Souza, C. G. M., Bracht, A., et al. (2018). Evaluation of diuron tolerance and biotransformation by the white-rot fungus Ganoderma lucidum. Fungal Biol. 122, 471–478. doi: 10.1016/j.funbio.2017.10.008
Daniel, G. (1994). Use of electron microscopy for aiding our understanding of wood biodegradation. FEMS Microbiol. Rev. 13, 199–233.
Daniel, G., and Nilsson, T. (1989). Interactions between soft rot fungi and CCA preservatives in Betula verrucosa. J. Inst.Wood Sci. 11, 162–171.
DeGroot, R., and Woodward, B. (1999). Using copper-tolerant fungi to biodegrade wood treated with copper based preservatives. Int. Biodeterior. Biodegrad. 44, 17–27. doi: 10.1016/S0964-8305(99)00047-5
Freeman, M. H., and McIntyre, C. R. (2008). A comprehensive review of copper-based wood preservatives. For. Prod. J. 58, 6–27.
Gadd, G. (1993). Tansley review no. 47: interactions of fungi with toxic metals. New Phytol. 124, 25–60. doi: 10.1111/j.1469-8137.1993.tb03796.x
Goni, R., Garcia, P., and Foissac, S. (2009). The qPCR data statistical analysis. Integromics S.L., 1–9.
Green, F., and Clausen, C. A. (2003). Copper tolerance of brown-rot fungi: time course of oxalic acid production. Int. Biodeterior. Biodegrad. 51, 145–149. doi: 10.1016/S0964-8305(02)00099-9
Green, F., and Clausen, C. A. (2005). Copper tolerance of brown-rot fungi: oxalic acid production in southern pine treated with arsenic-free preservatives. Int. Biodeterior. Biodegrad. 56, 75–79. doi: 10.1016/j.ibiod.2005.04.003
Gupta, A., and Lutsenko, S. (2012). Evolution of copper transporting ATPases in eukaryotic organisms. Curr. Genomics 13, 124–133. doi: 10.2174/138920212799860661
Hall, J. (2002). Cellular mechanisms for heavy metal detoxification and tolerance. J. Exp. Bot. 53, 1–11. doi: 10.1093/jexbot/53.366.1
Hastrup, A., Green, F., Clausen, C. A., and Jensen, B. (2005). Tolerance of Serpula lacrymans to copper-based wood preservatives. Int. Biodeterior. Biodegrad. 56, 173–177. doi: 10.1016/j.ibiod.2005.06.008
Hastrup, A., Jensen, B., Clausen, C. A., and Green, F. (2006). The effect of CaCl2 on growth rate, wood decay and oxalic acid accumulation in Serpula lacrymans and related brown-rot fungi. Holzforschung 60, 339–345. doi: 10.1515/HF.2006.054
Hastrup, A. C. S., Jensen, B., and Jellison, J. (2014). Fungal accumulation of metals from building materials during brown rot wood decay. Arch. Microbiol. 196, 565–574. doi: 10.1007/s00203-014-0993-z
Highley, T. L. (1973). Influence of carbon source on cellulase activity of white-rot and brown-rot fungi. Wood Fiber Sci. 5, 50–58.
Humar, M., Bucar, B., and Pohleven, F. (2006). Brown-rot decay of copper-impregnated wood. Int. Biodeterior. Biodegrad. 58, 9–14. doi: 10.1016/j.ibiod.2006.03.003
Humar, M., Petric, M., Pohleven, F., Sentjurc, M., and Kalan, P. (2002). Changes of EPR spectra of wood, impregnated with copper-based preservatives, during exposure to several wood rotting fungi. Holzforschung 56, 229–238. doi: 10.1515/HF.2002.038
Illman, B. L., Yang, V. W., and Ferge, L. (2000). Bioprocessing Preservative-Treated Waste Wood. IRG/WP 00-50145. The International Research Group on Wood Protection. Stockholm.
Ito, H., Inouhe, M., Tohoyama, H., and Joho, M. (2007). Characteristics of copper tolerance in Yarrowia lipolytica. Biometals 20, 773–780. doi: 10.1007/s10534-006-9040-0
Jarosz-Wilkolazka, A., and Graz, M. (2006). Organic acids production by white rot Basidiomycetes in the presence of metallic oxides. Can. J. Microbiol. 52, 779–785. doi: 10.1139/w06-032
Jaszek, M., Grzywnowicz, K., Malarczyk, E., and Leonowicz, A. (2006). Enhanced extra-cellular laccase activity as a part of the response system of white rot fungi: Trametes versicolor and Abortiporus biennis to paraquat-caused oxidative stress conditions. Pestic. Biochem. Physiol. 85, 147–154. doi: 10.1016/j.pestbp.2006.01.002
Jenkins, K. (2012). Evaluating the mechanism of oxalate synthesis of Fibroporia radiculosa isolates adapting to copper-tolerance. MS Thesis, Mississippi State University. 125.
Jenkins, K. M., Clausen, C. A., and Green, F. (2014). The effects of copper proximity on oxalate production in Fibroporia radiculosa. IRG/WP 14-10823. The International Research Group on Wood Protection. Stockholm.
Lattore, M., Olivares, F., Reyes-Jara, A., Lopez, G., and Gonzalez, M. (2011). CutC is induced late during copper exposure and can modify intracellular copper content in Enterococcus faecalis. Biochem. Biophys. Res. Commun. 406, 633–637. doi: 10.1016/j.bbrc.2011.02.109
Lebow, S. (2004). “Alternatives to chromated copper arsenate (CCA) for residential construction” in Proceedings of Environmental Impacts of Preservative-Treated Wood; February 8–11, 2004; Orlando, Florida, USA (Gainesville, Fla: Florida Center for Environmental Solutions).
Lebow, S. (2010). Wood Preservation. In: Wood Handbook: Wood as an Engineering Material. General Technical Report FPL-GTR-190. Department of Agriculture, Forest Service, Forest Products Laboratory. pp. 15.1–14.28. Madison, WI.
Lebow, S., Winandy, J., and Bender, D. (2004). Treated wood in transition: a look at CCA and the candidates to replace it. Wood Des. Focus, 3–8.
Lee, Y. P., Kim, S. H., Band, J. W., Lee, H. S., Kwak, S. S., and Kwon, S. Y. (2007). Enhanced tolerance to oxidative stress in transgenic tobacco plants expressing three antioxidant enzymes in chloroplasts. Plant Cell Rep. 26, 591–598. doi: 10.1007/s00299-006-0253-z
Leithoff, H., Stephan, I., Lenz, M. T., and Peek, R. D. (1995). Growth of the copper tolerant brown rot fungus Antrodia vaillantii on different substrates. IRG/WP 95-10121. The International Research Group on Wood Protection. Stockholm.
Li, J., Ji, C., Chen, Z., Yang, Z., Wang, Y., Fei, X., et al. (2005). Identification and characterization of a novel cut family cDNA that encodes human copper transporter protein CutC. Biochem. Biophys. Res. Commun. 337, 179–183. doi: 10.1016/j.bbrc.2005.09.029
Liew, F. J., and Schilling, J. S. (2012). Choice tests and neighbor effects during fungal brown rot of copper‐ and untreated wood. Int. Biodeterior. Biodegrad. 74, 7–10. doi: 10.1016/j.ibiod.2012.07.003
Livak, K., and Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maciel, G. M., Inacio, F. D., Sa-Nakanishi, A. B., Haminiuk, C. W. I., Castoldi, R., Comar, J. F., et al. (2013). Response of Ganoderma lucidum and Trametes sp. to the herbicide picloram: tolerance, antioxidants and production of ligninolytic enzymes. Pestic. Biochem. Physiol. 105, 84–92. doi: 10.1016/j.pestbp.2012.12.002
Magan, N., Fragoeiro, S., and Bastos, C. (2010). Environmental factors and bioremediation of xenobiotics using white rot fungi. Mycobiology 38, 238–248. doi: 10.4489/MYCO.2010.38.4.238
Migocka, M. (2015). Copper-transporting ATPases: the evolutionarily conserved machineries for balancing copper in living systems. Int. Union Biochem. Mol. Biol. 67, 737–745. doi: 10.1002/iub.1437
Murphy, R. J., and Levy, J. F. (1983). Production of copper oxalate by some copper tolerant fungi. Trans. Br. Mycol. Soc. 81, 165–168. doi: 10.1016/S0007-1536(83)80223-X
Noctor, G., and Foyer, C. H. (1998). Ascorbate and gluthathione: keeping active oxygen under control. Annu. Rev. Plant Biol. 49, 249–279. doi: 10.1146/annurev.arplant.49.1.249
Ohno, K. M., Clausen, C. A., Green, F., and Diehl, S. V. (2015). Insights into the mechanism of copper-tolerance in Fibroporia radiculosa: the biosynthesis of oxalate. Int. Biodeterior. Biodegrad. 105, 90–96. doi: 10.1016/j.ibiod.2015.08.016
Ohno, K. M., Kirker, G. T., Bishell, A. B., and Clausen, C. A. (2017). Untreated and copper-treated wood soaked in sodium oxalate: effects of decay by copper-tolerant and copper-sensitive fungi. IRG/WP 17-10888. The International Research Group on Wood Protection. Stockholm.
Pearce, D. A., and Sherman, F. (1999). Toxicity of copper, cobalt, and nickel salts is dependent on histidine metabolism in the yeast Saccharomyces cerevisiae. J. Bacteriol. 181, 4774–4779. doi: 10.1128/JB.181.16.4774-4779.1999
Peralta, R. M., Silva, B. P., Correa, R. C. G., Kato, C. G., Seixas, F. A. V., and Bracht, A. (2017). “Enzymes from basidiomycetes: peculiar and efficient tools for biotechnology” in Biotechnology of microbial enzymes: Production, biocatalysis and industrial applications. eds. G. Brahmachari, A. L. Demain, and J. L. Adrio (London: Academic Press), 119–149.
Pohleven, F., Breznikar, S., Kalan, P., and Petric, M. (1999). Determination of absorption, accumulation and transport of copper in mycelium of some wood decay fungi. IRG/WP 99-10323. The International Research Group on Wood Protection. Stockholm.
Pohleven, F., Humar, M., Amartey, S., and Benedik, J. (2002). Tolerance of wood decay fungi to commercial copper-based wood preservatives. IRG/WP 02-30291. The International Research Group on Wood Protection. Stockholm.
Presley, G. N., and Schilling, J. S. (2017). Distinct growth and secretome strategies for two taxonomically divergent brown rot fungi. Appl. Environ. Microbiol. 83, e02987–e03116. doi: 10.1128/AEM.02987-16
Raffa, N., Osherov, N., and Keller, N. P. (2019). Copper utilization, regulation, and acquisition by Aspergillus fumigatus. Int. J. Mol. Sci. 20:1980. doi: 10.3390/ijms20081980
Saitoh, Y., Izumitsu, K., and Tanaka, C. (2009). Phylogenetic analysis of heavy-metal ATPases in fungi and characterization of the copper-transporting ATPase of Cochliobolus heterostrophus. Mycol. Res. 113, 737–745. doi: 10.1016/j.mycres.2009.02.009
Schilling, J., and Inda, J. (2011). Assessing the relative bioavailability of copper to fungi degrading treated wood. Int. Biodeterior. Biodegrad. 65, 18–22. doi: 10.1016/j.ibiod.2010.05.013
Schilling, J., and Jellison, J. (2005). Oxalate regulation by two brown rot fungi decaying oxalate-amended and non-amended wood. Holzforschung 59, 681–688. doi: 10.1515/HF.2005.109
Schwartz, J. A., Olarte, K. T., Michalek, J. L., Jandu, G. S., Michel, S. L. J., and Bruno, V. M. (2013). Regulation of copper toxicity by Candida albicans GPA2. Eukaryot. Cell 12, 954–961. doi: 10.1128/EC.00344-12
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanisms in plants under stressful conditions. Aust. J. Bot. 2012, 1–26. doi: 10.1155/2012/217037
Shimanzono, H., and Takubo, K. (1952). The biochemistry of wood-destroying fungi: the Bavendamm’s reaction and the accumulation of oxalic acid by the wood-destroying fungi. Bull. Govern. Forest Exp.Stn. 53, 117–125.
Smith, A. D., Logeman, B. L., and Thiele, D. J. (2017). Copper acquisition and utilization in fungi. Annu. Rev. Microbiol. 71, 597–623. doi: 10.1146/annurev-micro-030117-020444
Somers, E. (1963). The uptake of copper by fungal cells. Ann. Appl. Biol. 51, 425–437. doi: 10.1111/j.1744-7348.1963.tb03710.x
Sutter, H. P., Jones, E. B. G., and Walchli, O. (1983). The mechanism of copper tolerance in Poria placenta (Fr.) Cke. And Poria vaillantii (Pers.). Fr. Mater. Org. 18, 241–262.
Sutter, H. P., Jones, E. B. G., and Walchli, O. (1984). Occurrence of crystalline hyphal sheath in Poria placenta (Fr.) Cke. J. Inst.Wood Sci. 10, 19–23.
Tamayo, E., Gomez-Gallego, T., Azcon-Aguilar, C., and Ferrol, N. (2014). Genome-wide analysis of copper, iron and zinc transporters in the arbuscular mycorrhizal fungus Rhizophagus irregularis. Front. Plant Sci. 5:547. doi: 10.3389/fpls.2014.00547
Tang, J. D., Parker, L. A., Perkins, A. D., Sonstegard, T. S., Schroeder, S. G., Nicholas, D. D., et al. (2013). Gene expression analysis of copper tolerance and wood decay in the brown rot fungus Fibroporia radiculosa. Appl. Environ. Microbiol. 79, 1523–1533. doi: 10.1128/AEM.02916-12
Tang, J., Perkins, A., Sonstegard, T., Schroeder, S., Burgess, S., and Diehl, S. (2012). Short-read sequencing for genomic analysis of the brown-rot fungus Fibroporia radiculosa. Appl. Environ. Microbiol. 78, 2272–2281. doi: 10.1128/AEM.06745-11
Tsunoda, K., Nagashimi, K., and Takahashi, M. (1997). High tolerance of wood-destroying brown-rot fungi to copper-based fungicides. Mater. Org. 31, 31–44.
Uldschmid, A., Dombi, R., and Marbach, K. (2003). Identification and functional expression of ctaA, a P-type ATPase gene involved in copper trafficking in Trametes versicolor. Microbiology 149, 2039–2048. doi: 10.1099/mic.0.26177-0
Weissman, Z., Berdicevsky, I., Cavari, B. Z., and Kornitzer, D. (2000). The high copper tolerance of Candida albicans is mediated by a P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 97, 3520–3525. doi: 10.1073/pnas.97.7.3520
Yahaya, Y. A., Don, M. M., and Bhatia, S. (2009). Biosorption of copper (II) onto immobilized cells of Pycnoporus sanguineus from aqueous solution: equilibrium and kinetic studies. J. Hazard. Mater. 161, 189–195. doi: 10.1016/j.jhazmat.2008.03.104
Zaefyzadeh, R. A., Quliyev, R. A., Babyeva, S. M., and Abbasov, M. A. (2009). The effect of the interaction between genotypes and drought stress on the superoxide dismutase and chlorophyll content in durum when landraces. Turk. J. Biol. 33, 1–7. doi: 10.3906/biy-0801-12
Zelinka, S. L., Jakes, J. E., Tang, J., Ohno, K., Bishell, A., Finney, L., et al. (2018). Fungal-copper interactions in wood examined with large field of view synchrotron-based X-ray fluorescence microscopy. Wood Mater. Sci. Eng. 14, 174–184. doi: 10.1080/17480272.2018.1458049
Keywords: copper-tolerance, brown-rot fungi, wood decay, gene expression, ATPases
Citation: Ohno KM, Bishell AB and Stanosz GR (2020) Gene Expression Analysis of Three Putative Copper-Transporting ATPases in Copper-Tolerant Fibroporia radiculosa. Front. Microbiol. 11:586940. doi: 10.3389/fmicb.2020.586940
Edited by:
Carlos Alberto Moreira-Filho, University of São Paulo, BrazilReviewed by:
Naresh Singhal, The University of Auckland, New ZealandSantosh Kr Karn, Sardar Bhagwan Singh Post Graduate Institute of Biomedical Science & Research, India
Copyright © 2020 Ohno, Bishell and Stanosz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katie M. Ohno, katie.m.ohno@usda.gov
 Katie M. Ohno
Katie M. Ohno Amy B. Bishell
Amy B. Bishell Glen R. Stanosz2
Glen R. Stanosz2