Abstract—
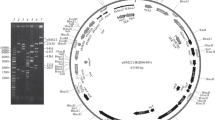
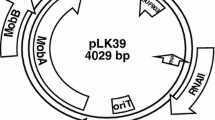
Analysis of the complete nucleotide sequence of the pBS72 plasmid from an environmental strain Bacillus subtilis 72 (102 254 bp) was used to characterize the genetic factors responsible for its vertical and horizontal gene transfer. Replicons similar to pBS72 were found to be present in B. subtilis isolated from various natural environments in Pakistan and the Netherlands (99% identity of the Rep proteins). The relaxase of the pBS72 plasmid, which is responsible for its conjugative transfer, was found to belong to the MobL family. A total of 142 open reading frames were revealed in the genome of the pBS72 plasmid, most of which (93) coded the synthesis of hypothetical polypeptides. Among the known proteins of the pBS72 plasmid genes in members of the genus Bacillus, the polypeptides were revealed, which were responsible for the interaction of bacterial cells with the environment, protection from foreign DNA and damage, as well as those involved in the cellular metabolism and carrying out regulatory functions. A system was developed in order to determine the role of the plasmid in activity of environmental and collection B. subtilis strains. In the presence of 13% NaCl, or at pH values of 4.5 and 11, survival of the environmental strain B. subtilis 72 and of the collection strain B. subtilis 168 was 4.33 and 13 times, 1.75 and 1.95 times, and 1.75 and 9.12 times higher, respectively. UV irradiation resulted in 2.25 times higher survival of B. subtilis 72 and 4.5 times higher survival of B. subtilis 168. The environmental strain B. subtilis 72 had more pronounced adaptive properties, which may indicate existence of additional genetic determinants in their chromosome; interaction of their products with the proteins of plasmid origin was probably responsible for higher resistance to increased osmolarity, high and low pH, and UV irradiation.





Similar content being viewed by others
REFERENCES
Anagnostopoulos, C. and Spizizen, J., Requirements for transformation in Bacillus subtilis,J. Bacteriol., 1961, vol. 81, pp. 741‒746.
Bron, S., Plasmids, in Molecular Biological Methods for Bacillus, Harwood, C.R. and Cutting, S.M., Eds., Chichester: John Wiley & Sons, 1990, pp. 75–174.
Bullock, W.O., Fernandez, J.M., and Short, J.M., XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection, BioTechniques, 1987, vol. 5, pp. 376–378.
Chambers, S.P., Prior, S.E., Barstow, D.A., and Minton, N.P., The pMTL nic-cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing, Gene, 1988, vol. 68, pp. 139‒149.
Dagan, T., Artzy-Randrup, Y., and Martin, W., Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution, Proc. Natl. Acad. Sci. USA, 2008, vol. 105, pp. 10039‒10044.
Edgar, R.C., Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 2010, vol. 26, pp. 2460‒2461.
Garcillán-Barcia, M.P., Francia, M.V., and de la Cruz, F., The diversity of conjugative relaxases and its application in plasmid classification, FEMS Microbiol. Rev., 2009, vol. 33, pp. 657‒687.
Guillen, C.J, Jones, G.H., Saldaña Gutiérrez, C., Hernández-Flores, J.L., Cruz Medina, J.A., Valenzuela Soto, J.H., Pacheco Hernández, S., Romero Gómez, S., and Morales Tlalpan, V., Critical minireview: the fate of tRNACys during oxidative stress in Bacillus subtilis,Biomolecules, 2017, vol. 7, art. 6. https://doi.org/10.3390/biom7010006
Harwood, C.R., Mouillon, J.M., Pohl, S., and Arnau, J., Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group, FEMS Microbiol. Rev., 2018, vol. 42, pp. 721‒738.
Kumar, S., Stecher, G., and Tamura, K., MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets, Mol. Biol. Evol., 2016, vol. 33, pp. 1870‒1874.
Lagodich, A.V., Cherva, E.A., Shtaniuk, Y.V., Prokulevich, V.A., Fomichev, Y.K., Prozorov, A.A., and Titok, M.A., Construction of a vector system for molecular cloning in Bacillus subtilis and Escherichia coli,Mol. Biol. (Moscow), 2005, vol. 39, pp. 306‒309.
Meijer, W.J., de Boer, A.J., van Tongeren, S., Venema, G., and Bron, S., Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon, Nucl. Acids Res., 1995, vol. 23, pp. 3214‒3223.
Meijer, W.J., Wisman, G.B., Terpstra, P., Thorsted, P.B., Thomas, C.M., Holsappel, S., Venema, G., and Bron, S., Rolling-circle plasmids from Bacillus subtilis: complete nucleotide sequences and analyses of genes of pTA1015, pTA1040, pTA1050 and pTA1060, and comparisons with related plasmids from gram-positive bacteria, FEMS Microbiol. Rev., 1998, vol. 21, pp. 337‒368.
Messer, W., The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication, FEMS Microbiol. Rev., 2002, vol. 26, pp. 355‒374.
Popa, O. and Dagan, T., Trends and barriers to lateral gene transfer in prokaryotes, Curr. Opin. Microbiol., 2011, vol. 14, pp. 615‒623.
Pluta, R., Boer, D.R., Lorenzo-Díaz, F., Russi, S., Gómez, H., Fernández-López, C., Pérez-Luque, R., Orozco, M., Espinosa, M., and Coll, M., Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance, Proc. Natl. Acad. Sci. USA, 2017, vol. 114. e: 6526‒6535.
Poluektova, E.U., Fedorina, E.A., Lotareva, O.V., and Prozorov, A.A., Plasmid transfer in bacilli by a self-transmissible plasmid p19 from a Bacillus subtilis soil strain, Plasmid, 2004, vol. 52, pp. 212‒217.
Ramachandran, G., Miguel-Arribas, A., Abia, D., Singh, P.K., Crespo, I., Gago-Córdoba, C., Hao, J.A., Luque-Ortega, J.R., Alfonso, C., Wu, L.J., Boer, D.R., and Meijer, W.J., Discovery of a new family of relaxases in Firmicutes bacteria, PLoS Genet., 2017, vol. 13. e: 1006586.
Rösch, T.C., Golman, W., Hucklesby, L., Gonzalez-Pastor, J.E., and Graumann, P.L., The presence of conjugative plasmid pLS20 affects global transcription of its Bacillus subtilis host and confers beneficial stress resistance to cells, Appl. Environ. Microbiol., 2014, vol. 80, pp. 1349–1358.
Sambrook, J., Fritsch, E., and Maniatis, T., Molecular Cloning: A Laboratory Manual, N.Y.: Cold Spring Harbor Laboratory, Cold Spring Harbor Publications, 1989, 2nd ed.
Shintani, M., Sanchez, Z.K., and Kimbara, K., Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy, Front. Microbiol., 2015, vol. 6, art. 242.
Smillie, C., Garcillán-Barcia, M.P., Francia, M.V., Rocha, E.P., and de la Cruz, F., Mobility of plasmids, Microbiol. Mol. Biol. Rev., 2010, vol. 74, pp. 434‒452.
Tanaka, T. and Ogura, M., A novel Bacillusnatto plasmid pLS32 capable of replication in Bacillus subtilis,FEBS Lett., 1998, vol. 422, pp. 243‒246.
te Riele, H., Michel, B., and Ehrlich, S.D., Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus,Proc. Natl. Acad. Sci. USA, 1986, vol. 83, pp. 2541‒2545.
Titok, M.A., Chapuis, J., Selezneva, Y.V., Lagodich, A.V., Prokulevich, V.A., Ehrlich, S.D., and Jannière, L., Bacillus subtilis soil isolates: plasmid replicon analysis and construction of a new theta-replicating vector, Plasmid, 2003, vol. 49, pp. 53‒62.
Weigel, C., Schmidt, A., Seitz, H., Tüngler, D., Welzeck, M., and Messer, W., The N-terminus promotes oligomerization of the Escherichia coli initiator protein DnaA, Mol. Microbiol., 1999, vol. 34, pp. 53‒66.
Yang, Y., Wu, H.J., Lin, L., Zhu, Q.Q., Borriss, R., and Gao, X.W., A plasmid-born Rap-Phr system regulates surfactin production, sporulation and genetic competence in the heterologous host, Bacillus subtilis OKB105, Appl. Microbiol. Biotechnol., 2015, vol. 99, pp. 7241‒7252.
Funding
This work was supported by the Belarussian Foundation for Basic Research, grant no. B18-070.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by D. Timchenko
Rights and permissions
About this article
Cite this article
Gurinovich, A.S., Titok, M.A. Molecular Genetic and Functional Analysis of the PBS72 Plasmid from Bacillus subtilis Environmental Isolates. Microbiology 89, 660–669 (2020). https://doi.org/10.1134/S0026261720060065
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261720060065