Abstract
Purpose of Review
Machine learning approaches—which seek to predict outcomes or classify patient features by recognizing patterns in large datasets—are increasingly applied to clinical epidemiology research on diabetes. Given its novelty and emergence in fields outside of biomedical research, machine learning terminology, techniques, and research findings may be unfamiliar to diabetes researchers. Our aim was to present the use of machine learning approaches in an approachable way, drawing from clinical epidemiological research in diabetes published from 1 Jan 2017 to 1 June 2020.
Recent Findings
Machine learning approaches using tree-based learners—which produce decision trees to help guide clinical interventions—frequently have higher sensitivity and specificity than traditional regression models for risk prediction. Machine learning approaches using neural networking and “deep learning” can be applied to medical image data, particularly for the identification and staging of diabetic retinopathy and skin ulcers. Among the machine learning approaches reviewed, researchers identified new strategies to develop standard datasets for rigorous comparisons across older and newer approaches, methods to illustrate how a machine learner was treating underlying data, and approaches to improve the transparency of the machine learning process.
Summary
Machine learning approaches have the potential to improve risk stratification and outcome prediction for clinical epidemiology applications. Achieving this potential would be facilitated by use of universal open-source datasets for fair comparisons. More work remains in the application of strategies to communicate how the machine learners are generating their predictions.





Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Doupe P, Faghmous J, Basu S. Machine learning for health services researchers. Value Heal. 2019.
Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. 2nd edition. New York: Springer; 2011. A core textbook to learn essential machine learning principles.
Doupe P, Faghmous J, Basu S. Machine learning for health services researchers. Value Heal. 2019;22:808–15.
Basu S, Faghmous JH, Doupe P. Machine learning methods for precision medicine research designed to reduce health disparities: a structured tutorial. Ethn Dis. 2020;30:217–28.
Sutton RS, Barto AG. Reinforcement learning: an introduction. MIT Press; 2018.
Breiman L. Random forests. Mach Learn. 2001 Oct 1;45(1):5–32.
Friedman JH. Stochastic gradient boosting. Comput Stat Data Anal. 2002;38:367–78.
Carin L, Pencina MJ. On deep learning for medical image analysis. JAMA. 2018.
Ngiam J, Khosla A, Kim M, Nam J, Lee H, Ng AY. Multimodal deep learning: International Conference on Machine Learning; 2011 Jan 1.
Chowdhury MZ, Yeasmin F, Rabi DM, Ronksley PE, Turin TC. Predicting the risk of stroke among patients with type 2 diabetes: a systematic review and metaanalysis of C-statistics. BMJ Open. 2019 Aug 1;9(8):e025579.
Mueller L, Berhanu P, Bouchard J, Alas V, Elder K, Thai N, et al. Application of machine learning models to evaluate hypoglycemia risk in type 2 diabetes. Diabetes Ther Res Treat Educ Diabetes Relat Disord. 2020;11:681–99.
Wittenbecher C, Štambuk T, Kuxhaus O, Rudman N, Vučković F, Štambuk J, et al. Plasma N-glycans as emerging biomarkers of cardiometabolic risk: a prospective investigation intheepic-potsdamcohortstudy. Diabetes Care [Internet]. Department of Molecular Epidemiology, German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal, Germany; 2020;43:661–8. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85081141185&doi=10.2337%2Fdc19-1507&partnerID=40&md5=441a1d1dce973452b7387c501b06e766NS.
Balani J, Hyer SL, Shehata H, Mohareb F. Visceral fat mass as a novel risk factor for predicting gestational diabetes in obese pregnant women. Obstet Med [Internet]. Department of Endocrinology, Epsom and St Helier University Hospitals NHS Trust, Surrey, United Kingdom; 2018;11:121–5. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85044067988&doi=10.1177%2F1753495X17754149&partnerID=40&md5=df8a39f1569e2a649d9aa12b834477c7. Accessed 19 July 2020.
Rawshani A, Sattar N, Franzén S, Mcguire DK, Eliasson B, Svensson A-M, et al. Relative prognostic importance and optimal levels of risk factors for mortality and cardiovascular outcomes in type 1 diabetes mellitus. Circulation [Internet]. Department of Molecular and Clinical Medicine, Institute of Medicine, University of Gothenburg, Sweden; 2019;139:1900–12. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85064934502&doi=10.1161%2FCIRCULATIONAHA.118.037454&partnerID=40&md5=5f052f741eb86bc729764e1ce40b7593. Accessed 19 July 2020.
Ye L, Lee T-S, Chi R. A hybrid machine learning scheme to analyze the risk factors of breast Cancer outcome in patients with diabetes mellitus. J Univers Comput Sci [internet]. Inffeldgasse 16C, GRAZ, A-8010, Austria: Graz Univ Technolgoy, Inst Information Systems COMPUTER Media-IICM; 2018;24:665–81. Accessed 19 July 2020.
Choi BG, Rha SW, Kim SW, Kang JH, Park JY, Noh YK. Machine learning for the prediction of new-onset diabetes mellitus during 5-year follow-up in non-diabetic patients with cardiovascular risks. Yonsei Med J. 2019 Feb 1;60(2):191–9.
Ferreira ACBH, Ferreira DD, Oliveira HC, Resende ICD, Anjos A, Lopes MHBDM. Competitive neural layer-based method to identify people with high risk for diabetic foot. Comput Biol Med [Internet]. School of Nursing, Universidade Estadual de Campinas, Campinas, São Paulo, Brazil; 2020;120. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85082864958&doi=10.1016%2Fj.compbiomed.2020.103744&partnerID=40&md5=aaebc630d2238db60f48c2576fff2594.
Talaei-Khoei A, Wilson JM. Identifying people at risk of developing type 2 diabetes: a comparison of predictive analytics techniques and predictor variables. Int J Med Inform [Internet]. Department of Information Systems, Ansari College of Business, University of Nevada, Reno, United States; 2018;119:22–38. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85053074292&doi=10.1016%2Fj.ijmedinf.2018.08.008&partnerID=40&md5=e960f942d99f9f1afbf8224d4a9b82cc. Accessed 19 July 2020.
Sarda A, Munuswamy S, Sarda S, Subramanian V. Using passive smartphone sensing for improved risk stratification of patients with depression and diabetes: cross-sectional observational study. JMIR mHealth uHealth [Internet]. Sarda Centre for Diabetes and Selfcare, Aurangabad, India; 2019;7. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85072962699&doi=10.2196%2F11041&partnerID=40&md5=cf0a44b2af5e385973752105501a2585. Accessed 19 July 2020.
Zarkogianni K, Athanasiou M, Thanopoulou AC. Comparison of machine learning approaches toward assessing the risk of developing cardiovascular disease as a Long-term diabetes complication. IEEE J Biomed Heal Inform. 2018;22:1637–47. Accessed 19 July 2020.
Basu S, Raghavan S, Wexler DJ, Berkowitz SA. Characteristics associated with decreased or increased mortality risk from glycemic therapy among patients with type 2 diabetes and high cardiovascular risk: machine learning analysis of the ACCORD trial. Diabetes Care United States. 2018;41:604–12.
Aminian A, Zajichek A, Arterburn DE, Wolski KE, Brethauer SA, Schauer PR, et al. Predicting 10-year risk of end-organ complications of type 2 diabetes with and without metabolic surgery: a machine learning approach. Diabetes Care. 2020;43:852–9.
Oviedo S, Contreras I, Quirós C, Giménez M, Conget I, Vehi J. Risk-based postprandial hypoglycemia forecasting using supervised learning. Int J Med Inform [Internet]. Institut d’Informatica i Aplicacions. Universitat de Girona, Spain; 2019;126:1–8. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85062808679&doi=10.1016%2Fj.ijmedinf.2019.03.008&partnerID=40&md5=eceb673ea4fbabe36a4afba2649cc188.
Bernardini M, Morettini M, Romeo L, Frontoni E, Burattini L. Early temporal prediction of type 2 diabetes risk condition from a general practitioner electronic health record: a multiple instance boosting approach. Artif Intell Med [Internet]. Department of Information Engineering (DII), Università Politecnica delle Marche, Ancona, Italy; 2020;105. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85084579225&doi=10.1016%2Fj.artmed.2020.101847&partnerID=40&md5=db4c6fc2646ad6540af29321b95f7685. Accessed 19 July 2020.
Hertroijs DFL, Elissen AMJ, Brouwers MCGJ, Schaper NC, Köhler S, Popa MC, et al. A risk score including body mass index, glycated haemoglobin and triglycerides predicts future glycaemic control in people with type 2 diabetes. Diabetes, Obes Metab [Internet]. Department of Health Services Research, Care and Public Health Research Institute, Faculty of Health, Medicine and Life Sciences, Maastricht University, Maastricht, Netherlands; 2018;20:681–8. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85035047296&doi=10.1111%2Fdom.13148&partnerID=40&md5=cc25caa1b18492b53db57b89048fda00. Accessed 19 July 2020.
Solodskikh SA, Velikorondy AS, Popov VN. Predictive estimates of risks associated with type 2 diabetes mellitus on the basis of biochemical biomarkers and derived time-dependent parameters. J Comput Biol [Internet]. Department of Genetics, Cytology and Bioengineering, Voronezh State University, Universitetskaya sq 1, Voronezh, 394000, Russian Federation; 2019;26:1041–9. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85073087062&doi=10.1089%2Fcmb.2019.0028&partnerID=40&md5=46fb5fcb001774dee3184ce96df89377. Accessed 19 July 2020.
Farran B, AlWotayan R, Alkandari H, Al-Abdulrazzaq D, Channanath A, Thanaraj TA. Use of non-invasive parameters and machine-learning algorithms for predicting future risk of type 2 diabetes: a retrospective cohort study of health data from Kuwait. Front Endocrinol (Lausanne). 2019;10:624. Accessed 19 July 2020.
Frohnert BI, Webb-Robertson B-J, Bramer LM, Reehl SM, Waugh K, Steck AK, et al. Predictive modeling of type 1 diabetes stages using disparate data sources. Diabetes [Internet]. Barbara Davis Center for Diabetes, School of Medicine, University of Colorado, Aurora, CO, United States; 2020;69:238–48. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85078380011&doi=10.2337%2Fdb18-1263&partnerID=40&md5=4b8e78193c87a2631389ba191f267853.
Bhat V, Tazari M, Watt KD, Bhat M. New-onset diabetes and preexisting diabetes are associated with comparable reduction in Long-term survival after liver transplant: a machine learning approach. Mayo Clin Proc. 2018;93:1794–802 Accessed 19 July 2020..
Neugebauer R, Schroeder EB, Reynolds K, Schmittdiel JA, Loes L, Dyer W, et al. Comparison of mortality and major cardiovascular events among adults with type 2 diabetes using human vs analogue insulins. JAMA Netw open [Internet]. Division of Research, Kaiser Permanente Northern California, Oakland, United States; 2020;3:e1918554–e1918554. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85078309632&doi=10.1001%2Fjamanetworkopen.2019.18554&partnerID=40&md5=10f95c843c93eb333d6dad15905d3bc3.
Yamada T, Iwasaki K, Maedera S, Ito K, Takeshima T, Noma H, et al. Myocardial infarction in type 2 diabetes using sodium-glucose co-transporter-2 inhibitors, dipeptidyl peptidase-4 inhibitors or glucagon-like peptide-1 receptor agonists: proportional hazards analysis by deep neural network based machine learning. Curr Med Res Opin. 2020;36:403–9. Accessed 19 July 2020.
Garcia-Carretero R, Vigil-Medina L, Barquero-Perez O, Ramos-Lopez J. Pulse wave velocity and machine learning to predict cardiovascular outcomes in prediabetic and diabetic populations. J Med Syst [Internet]. Department of Internal Medicine, Mostoles University Hospital, Rey Juan Carlos University, Móstoles, Spain; 2020;44. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85076293329&doi=10.1007%2Fs10916-019-1479-y&partnerID=40&md5=8f6d29e5eb5815f35ab4bf4cf2e70e07.
Hegde H, Shimpi N, Panny A, Glurich I, Christie P, Acharya A. Development of non-invasive diabetes risk prediction models as decision support tools designed for application in the dental clinical environment. Informatics Med Unlocked [Internet]. Center for Oral and Systemic Health, Marshfield Clinic Research Institute, Marshfield, WI, United States; 2019;17. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85074276148&doi=10.1016%2Fj.imu.2019.100254&partnerID=40&md5=6c8c8d674808fe861b64d0a1d858ee89. Accessed 19 July 2020.
Dankwa-Mullan I, Rivo M, Sepulveda M, Park Y, Snowdon J, Rhee K. Transforming diabetes care through artificial intelligence: the future is here. Popul Health Manag. 2019;22:229–42. Accessed 19 July 2020.
Lafta R, Zhang J, Tao X, Zhu X, Li H, Chang L, et al. A general extensible learning approach for multi-disease recommendations in a telehealth environment. Pattern Recognit Lett [Internet]. Faculty of Health, Engineering and Sciences, University of Southern Queensland, Australia; 2020;132:106–14. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85056445709&doi=10.1016%2Fj.patrec.2018.11.006&partnerID=40&md5=97aa97ad3eb17b0cf8d5d858b440a30f.
Dzakiyullah NR, Burhanuddin MA, Ikram RRR, Ghani KA, Setyonugroho W. Machine learning methods for diabetes prediction. Int J Innov Technol Explor Eng [Internet]. Faculty of Information and Communication Technology, Universiti Teknikal Malaysia Melaka (UTeM), Melaka, Malaysia; 2019;8:2199–205. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85073722910&doi=10.35940%2Fijitee.L2973.1081219&partnerID=40&md5=ecb9ae939a13a83001fed70fe6c3cc97. Accessed 19 July 2020.
Cahn A, Shoshan A, Sagiv T, Yesharim R, Goshen R, Shalev V, et al. Prediction of progression from pre-diabetes to diabetes: development and validation of a machine learning model. Diabetes Metab Res Rev. 2020;36:e3252. Accessed 19 July 2020.
Islam MM, Rahman MJ, Chandra Roy D, Maniruzzaman M. Automated detection and classification of diabetes disease based on Bangladesh demographic and health survey data, 2011 using machine learning approach. Diabetes Metab Syndr. 2020;14:217–9.
Ganguli M, Beer JC, Zmuda JM, Ryan CM, Sullivan KJ, Chang C-CH, et al. Aging, diabetes, obesity, and cognitive decline: a population-based study. J Am Geriatr Soc [Internet]. Department of Psychiatry, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States; 2020;68:991–8. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85079041449&doi=10.1111%2Fjgs.16321&partnerID=40&md5=d253ea7014177ca0dfdd5cb380899b5c.
Peddinti G, Cobb J, Yengo L, Froguel P, Kravić J, Balkau B, et al. Early metabolic markers identify potential targets for the prevention of type 2 diabetes. Diabetologia [Internet]. Institute for Molecular Medicine Finland (FIMM), Nordic EMBL Partnership for Molecular Medicine, University of Helsinki, Helsinki, Finland; 2017;60:1740–50. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85020459515&doi=10.1007%2Fs00125-017-4325-0&partnerID=40&md5=d9f70ce7bfcc0f87248556e538ed23b7. Accessed 19 July 2020.
Cooray SD, Boyle JA, Soldatos G, Wijeyaratne LA, Teede HJ. Prognostic prediction models for pregnancy complications in women with gestational diabetes: a protocol for systematic review, critical appraisal and meta-analysis. Syst Rev. 2019;8:270. Accessed 19 July 2020.
Song X, Waitman LR, Hu Y, Yu ASL, Robins D, Liu M. Robust clinical marker identification for diabetic kidney disease with ensemble feature selection. J Am Med Informatics Assoc [Internet]. Department of Internal Medicine, Division of Medical Informatics, University of Kansas Medical Center, 3001B Stud. Center, 3901 Rainbow Boulevard, Kansas City, KS 66160, United States; 2019;26:242–53. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85060803404&doi=10.1093%2Fjamia%2Focy165&partnerID=40&md5=acb29a6cabc7d95810cd4ccab09c1363.
Chowdhury MZI, Yeasmin F, Rabi DM, Ronksley PE, Turin TC. Prognostic tools for cardiovascular disease in patients with type 2 diabetes: a systematic review and meta-analysis of C-statistics. J Diabetes Complicat. 2019;33:98–111. Accessed 19 July 2020.
Liu Y, Ye S, Xiao X, Sun C, Wang G, Wang G, et al. Machine learning for tuning, selection, and ensemble of multiple risk scores for predicting type 2 diabetes. Risk Manag Healthc Policy. 2019;12:189–98.
Vehí J, Contreras I, Oviedo S, Biagi L, Bertachi A. Prediction and prevention of hypoglycaemic events in type-1 diabetic patients using machine learning. Health Informatics J [Internet]. Universitat de Girona, Spain; 2020;26:703–18. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85067878120&doi=10.1177%2F1460458219850682&partnerID=40&md5=2e7215cfe864523d3363e61424d30524.
Khan SR, Mohan H, Liu Y, Batchuluun B, Gohil H, Al Rijjal D, et al. The discovery of novel predictive biomarkers and early-stage pathophysiology for the transition from gestational diabetes to type 2 diabetes. Diabetologia [Internet]. Endocrine and Diabetes Platform, Department of Physiology, University of Toronto, Medical Sciences Building, Room 3352, 1 King’s College Circle, Toronto, ON M5S 1A8, Canada; 2019;62:687–703. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85060160755&doi=10.1007%2Fs00125-018-4800-2&partnerID=40&md5=313eb7e3300591dc963203a9a478b9f8. Accessed 19 July 2020.
Ravaut M, Sadeghi H, Leung KK, Volkovs M, Rosella LC. Diabetes mellitus forecasting using population health data in Ontario, Canada. 2019;1–18. Available from: http://arxiv.org/abs/1904.04137. Accessed 19 July 2020.
Nowak C, Carlsson AC, Östgren CJ, Nyström FH, Alam M, Feldreich T, et al. Multiplex proteomics for prediction of major cardiovascular events in type 2 diabetes. Diabetologia [Internet]. Division of Family Medicine and Primary Care, Department of Neurobiology, Care Sciences and Society (NVS), Karolinska Institutet, Alfred Nobels Allé 23, Huddinge, SE 14183, Sweden; 2018;61:1748–57. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85047249027&doi=10.1007%2Fs00125-018-4641-z&partnerID=40&md5=3d1e7cc928b00590060690ba33e6fcf4. Accessed 19 July 2020.
Cao P, Ren F, Wan C, Yang J, Zaiane O. Efficient multi-kernel multi-instance learning using weakly supervised and imbalanced data for diabetic retinopathy diagnosis. Comput Med Imaging Graph [Internet]. Computer Science and Engineering, Northeastern University, Shenyang, China; 2018;69:112–24. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85053423136&doi=10.1016%2Fj.compmedimag.2018.08.008&partnerID=40&md5=9287ddf72eccdc4e305cc6ac1ac62e7c. Accessed 19 July 2020.
Pettus J, Roussel R, Liz Zhou F, Bosnyak Z, Westerbacka J, Berria R, et al. Rates of hypoglycemia predicted in patients with type 2 diabetes on insulin glargine 300 u/ml versus first- and second-generation basal insulin analogs: the real-world lightning study. Diabetes Ther [Internet]. School of Medicine, University of California, San Diego, CA, United States; 2019;10:617–33. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85065769762&doi=10.1007%2Fs13300-019-0568-8&partnerID=40&md5=0e47a294bb98fcb72575a4f71dc9d9b7.
Saleh E, Błaszczyński J, Moreno A, Valls A, Romero-Aroca P, de la Riva-Fernández S, et al. Learning ensemble classifiers for diabetic retinopathy assessment. Artif Intell Med [Internet]. Departament d’Enginyeria Informàtica i Matemàtiques, Universitat Rovira i Virgili, Tarragona, Spain; 2018;85:50–63. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85030629171&doi=10.1016%2Fj.artmed.2017.09.006&partnerID=40&md5=e95a962380b954459e3063de94d13caf. Accessed 19 July 2020.
Cappon G, Vettoretti M, Marturano F, Facchinetti A, Sparacino G. A neural-network-based approach to personalize insulin bolus calculation using continuous glucose monitoring. J Diabetes Sci Technol [Internet]. Department of Information Engineering, University of Padova, Padova, PD, Italy; 2018;12:265–72. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85042874905&doi=10.1177%2F1932296818759558&partnerID=40&md5=e39c3b96dddb3ef53fd05872272a237b. Accessed 19 July 2020.
Singh N, Singh P, Bhagat D. A rule extraction approach from support vector machines for diagnosing hypertension among diabetics. Expert Syst Appl [Internet]. Department of Computer Science and Engineering, National Institute of Technology, Raipur, Chhattisgarh 492001, India; 2019;130:188–205. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85064536804&doi=10.1016%2Fj.eswa.2019.04.029&partnerID=40&md5=069d2672dfcb50ae896a34b7610f5512. Accessed 19 July 2020.
Wang J, Wang M-Y, Wang H, Liu H-W, Lu R, Duan T-Q, et al. Status of glycosylated hemoglobin and prediction of glycemic control among patients with insulin-treated type 2 diabetes in North China: a multicenter observational study. Chin Med J (Engl) [Internet]. Department of Health Statistics, College of Public Health, Tianjin Medical University, No. 22 Qixiangtai Road, Heping District, Tianjin, 300070, China; 2020;133:17–24. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85077765228&doi=10.1097%2FCM9.0000000000000585&partnerID=40&md5=119863b171e2f6b9a4583b01328b05ce. Accessed 19 July 2020.
Segar MW, Vaduganathan M, Patel KV, McGuire DK, Butler J, Fonarow GC, et al. Machine learning to predict the risk of incident heart failure hospitalization among patients with diabetes: the WATCH-DM risk score. Diabetes Care. 2019;42:2298–306. Accessed 19 July 2020.
Nielsen KB, Lautrup ML, Andersen JKH, Savarimuthu TR, Grauslund J. Deep learning-based algorithms in screening of diabetic retinopathy: a systematic review of diagnostic performance. Ophthalmol Retin. 2019;3:294–304.
Nusinovici S, Tham YC, Chak Yan MY, Wei Ting DS, Li J, Sabanayagam C, et al. Logistic regression was as good as machine learning for predicting major chronic diseases. J Clin Epidemiol [Internet]. Singapore Eye Research Institute, Singapore National Eye Centre, Singapore, Singapore; 2020;122:56–69. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85082738590&doi=10.1016%2Fj.jclinepi.2020.03.002&partnerID=40&md5=4ccd31e001da804dc9b1781ca1003e6d.
Lai H, Huang H, Keshavjee K, Guergachi A, Gao X. Predictive models for diabetes mellitus using machine learning techniques. BMC Endocr Disord. 2019;19:101. Accessed 19 July 2020.
van der Heijden AA, Nijpels G, Badloe F, Lovejoy HL, Peelen LM, Feenstra TL, et al. Prediction models for development of retinopathy in people with type 2 diabetes: systematic review and external validation in a Dutch primary care setting. Diabetologia. 2020;63:1110–9
Vu GT, Tran BX, McIntyre RS, Pham HQ, Phan HT, Ha GH, et al. Modeling the research landscapes of artificial intelligence applications in diabetes (GAPresearch). Int J Environ Res Public Health [Internet]. Center of Excellence in Evidence-based Medicine, Nguyen Tat Thanh University, Ho Chi Minh City, 700000, Viet Nam; 2020;17. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85082053323&doi=10.3390%2Fijerph17061982&partnerID=40&md5=61a3b606e68edef55f28144d01213785.
Tsao H-Y, Chan P-Y, Su EC-Y. Predicting diabetic retinopathy and identifying interpretable biomedical features using machine learning algorithms. BMC Bioinformatics [Internet]. Taipei Medical University, Graduate Institute of Biomedical Informatics, College of Medical Science and Technology, Taipei, 106, Taiwan; 2018;19. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85051590520&doi=10.1186%2Fs12859-018-2277-0&partnerID=40&md5=542f1f3f5ff4b2263a4689795b0cd38b. Accessed 19 July 2020.
Dinh A, Miertschin S, Young A, Mohanty SD. A data-driven approach to predicting diabetes and cardiovascular disease with machine learning. BMC Med Inform Decis Mak. 2019;19:211. Accessed 19 July 2020.
Ali RE, El-Kadi H, Labib SS, Saad YI. Prediction of potential-diabetic obese-patients using machine learning techniques. Int J Adv Comput Sci Appl [Internet]. Faculty of Computer Science, MSA University, Faculty of Computers and Artificial Intelligence, Giza, Egypt; 2019;10:80–8. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85072292817&partnerID=40&md5=e99112fcf1e63faa8b4d8a808faf5076.
Garcia-Carretero R, Vigil-Medina L, Mora-Jimenez I, Soguero-Ruiz C, Barquero-Perez O, Ramos-Lopez J. Use of a K-nearest neighbors model to predict the development of type 2 diabetes within 2 years in an obese, hypertensive population. Med Biol Eng Comput [Internet]. Internal Medicine Department, Mostoles University Hospital, Rey Juan Carlos University, Calle Rio Jucar, s/n, 28935, Mostoles (Madrid), Spain; 2020;58:991–1002. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85080065490&doi=10.1007%2Fs11517-020-02132-w&partnerID=40&md5=7e8f44664d4a5397416263f4e0aa7bdd. Accessed 19 July 2020.
Basu S, Narayanaswamy R. A prediction model for uncontrolled type 2 diabetes mellitus incorporating area-level social determinants of health. Med Care United States. 2019;57:592–600. Accessed 19 July 2020.
Xie Z, Nikolayeva O, Luo J, Li D. Building risk prediction models for type 2 diabetes using machine learning techniques. Prev Chronic Dis. 2019;16:E130.
Thirunavukkarasu U, Umapathy S, Krishnan PT, Janardanan K. Human tongue thermography could be a prognostic tool for prescreening the type II diabetes mellitus. Evidence-based Complement Altern Med [Internet]. Department of Biomedical Engineering, SRM Institute of Science and Technology, Kattankulathur, Tamil Nadu, 603203, India; 2020;2020. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85078657656&doi=10.1155%2F2020%2F3186208&partnerID=40&md5=ab1684feda30611b6ba790f20f15a332.
McCoy RG, Ngufor C, Van Houten HK, Caffo B, Shah ND. Trajectories of glycemic change in a national cohort of adults with previously controlled type 2 diabetes. Med Care [Internet]. Department of Medicine, Division of Primary Care Internal Medicine, Mayo Clinic, 200 First Street SW, Rochester, MN 55905, United States; 2017;55:956–64. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85031817336&doi=10.1097%2FMLR.0000000000000807&partnerID=40&md5=dbbd747981ad7efa6c7755c19ec9a688. Accessed 19 July 2020.
Goyal M, Reeves ND, Rajbhandari S, Ahmad N. Recognition of ischaemia and infection in diabetic foot ulcers: dataset and techniques. Accessed 19 July 2020.
Woldaregay AZ, Årsand E, Botsis T, Albers D, Mamykina L, Hartvigsen G. Datadriven blood glucose pattern classification and anomalies detection: machine learning applications in type 1 diabetes. J Med Internet Res. 2019;21(5):e11030.
Zheng T, Ye W, Wang X, Li X, Zhang J, Little J, et al. A simple model to predict risk of gestational diabetes mellitus from 8 to 20 weeks of gestation in Chinese women. BMC Pregnancy Childbirth [Internet]. Obstetric and Gynecology Department, Xinhua Hospital, Shanghai Jiao Tong University, School of Medicine, Shanghai, China; 2019;19. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85069467377&doi=10.1186%2Fs12884-019-2374-8&partnerID=40&md5=ce3295e1494d3360429cf71c0e39a837.
López B, Torrent-Fontbona F, Viñas R, Fernández-Real JM. Single nucleotide polymorphism relevance learning with random forests for type 2 diabetes risk prediction. Artif Intell Med. 2018;85:43–9. Accessed 19 July 2020.
Wu X-W, Yang H-B, Yuan R, Long E-W, Tong R-S. Predictive models of medication non-adherence risks of patients with T2D based on multiple machine learning algorithms. BMJ Open Diabetes Res Care [Internet]. Personalized Drug Therapy Key Laboratory of Sichuan Province, School of Medicine, University of Electronic Science and Technology of China, Chengdu, China; 2020;8. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85081599410&doi=10.1136%2Fbmjdrc-2019-001055&partnerID=40&md5=bd4a99ee03795b3186911213e24a29d6.
Basu S, Sussman JB, Berkowitz SA, Hayward RA, Yudkin JS. Development and validation of risk equations for complications of type 2 diabetes (RECODe) using individual participant data from randomised trials. Lancet Diabetes Endocrinol England. 2017;5:788–98. Accessed 19 July 2020.
Duan T, Rajpurkar P, Laird D, Ng AY, Basu S. Clinical value of predicting individual treatment effects for intensive blood pressure therapy. Circ Cardiovasc Qual Outcomes United States. 2019;12:e005010.
Basu S, Sussman JB, Berkowitz SA, Hayward RA, Bertoni AG, Correa A, et al. Validation of risk equations for complications of type 2 diabetes (RECODe) using individual participant data from diverse longitudinal cohorts in the U.S. Diabetes Care United States. 2018;41:586–95.
van Klaveren D, Steyerberg EW, Serruys PW, Kent DM. The proposed ‘concordance-statistic for benefit’ provided a useful metric when modeling heterogeneous treatment effects. J Clin Epidemiol. 2018;94:59–68.
Jung LC, Wang H, Li X, Wu C. A machine learning method for selection of genetic variants to increase prediction accuracy of type 2 diabetes mellitus using sequencing data. Stat Anal Data Min. 2020;13:261–81 111 River St, Hoboken 07030-5774, NJ USA: Wiley;.
Nguyen BP, Pham HN, Tran H, Nghiem N, Nguyen QH, Do TTT, et al. Predicting the onset of type 2 diabetes using wide and deep learning with electronic health records. Comput Methods Prog Biomed. 2019;182:105055.
De Silva K, Jönsson D, Demmer RT. A combined strategy of feature selection and machine learning to identify predictors of prediabetes. J Am Med Informatics Assoc [Internet]. Department of Clinical Sciences, Faculty of Medicine, Lund University, Lund, Sweden; 2020;27:396–406. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85079353320&doi=10.1093%2Fjamia%2Focz204&partnerID=40&md5=c9c8c99600bab772a69c2a31581a3510.
Tama BA, Rhee K-H. Tree-based classifier ensembles for early detection method of diabetes: an exploratory study. Artif Intell Rev [Internet]. IT Convergence and Application Engineering, Pukyong National University, (48513) Daeyon Campus, 45, Yongso-ro, Nam-Gu, Busan, South Korea; 2019;51:355–70. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85019655672&doi=10.1007%2Fs10462-017-9565-3&partnerID=40&md5=346269e280f7ea4ccd2557d236431c85. Accessed 19 July 2020.
Orlando JI, Prokofyeva E, del Fresno M, Blaschko MB. An ensemble deep learning based approach for red lesion detection in fundus images. Comput Methods Prog Biomed. 2018;153:115–27. Accessed 19 July 2020.
Akula R, Nguyen N, Garibay I. Supervised machine learning based ensemble model for accurate prediction of type 2 diabetes. 2018;.
Murphree DH, Arabmakki E, Ngufor C, Storlie CB, McCoy RG. Stacked classifiers for individualized prediction of glycemic control following initiation of metformin therapy in type 2 diabetes. Comput Biol Med. 2018 Dec 1;103:109–15.
Ma L, Zheng J. Single-cell gene expression analysis reveals β-cell dysfunction and deficit mechanisms in type 2 diabetes. BMC Bioinform. 2018;19:515. Accessed 19 July 2020.
Li L, Krznar P, Erban A, Agazzi A, Martin-Levilain J, Supale S, et al. Metabolomics identifies a biomarker revealing in vivo loss of functional β-cell mass before diabetes onset. Diabetes. 2019.
Hathaway QA, Roth SM, Pinti MV, Sprando DC, Kunovac A, Durr AJ, et al. Machine-learning to stratify diabetic patients using novel cardiac biomarkers and integrative genomics. Cardiovasc Diabetol. 2019;18:78. Accessed 19 July 2020.
Liu ZP, Gao R. Detecting pathway biomarkers of diabetic progression with differential entropy. J Biomed Inform. 2018;82:143–53.
Bernardini M, Morettini M, Romeo L, Frontoni E, Burattini L. TyG-er: an ensemble regression Forest approach for identification of clinical factors related to insulin resistance condition using electronic health records. Comput Biol Med. 2019;112:103358.
Khan SR, Mohan H, Liu Y, Batchuluun B, Gohil H, Al Rijjal D, et al. The discovery of novel predictive biomarkers and early-stage pathophysiology for the transition from gestational diabetes to type 2 diabetes. Diabetologia. 2019.
Wittenbecher C, Štambuk T, Kuxhaus O, Rudman N, Vučković F, Štambuk J, et al. Plasma N-glycans as emerging biomarkers of cardiometabolic risk: a prospective investigation in the EPIC-potsdam cohort study. Diabetes Care. 2020 Mar 1;43(3):661–8.
Guo Z, Yang F, Zhang J, Zhang Z, Li K, Tian Q, et al. Whole-genome promoter profiling of plasma DNA exhibits diagnostic value for placenta-origin pregnancy complications. Adv Sci. 2020.
Ding L, Fan L, Xu X, Fu J, Xue Y. Identification of core genes and pathways in type 2 diabetes mellitus by bioinformatics analysis. Mol Med Rep. 2019 Sep 1;20(3):2597–608.
Jung LC, Wang H, Li X, Wu C. A machine learning method for selection of genetic variants to increase prediction accuracy of type 2 diabetes mellitus using sequencing data. Stat Anal Data Min 2020.
Dong SS, Guo Y, Yao S, Chen YX, He MN, Zhang YJ, et al. Integrating regulatory features data for prediction of functional disease-associated SNPs. Brief Bioinform. 2019.
Han W, Ye Y. A repository of microbial marker genes related to human health and diseases for host phenotype prediction using microbiome data: Pacific Symposium on Biocomputing; 2019 Jan 1. p. 236–47.
Dong SS, Guo Y, Yao S, Chen YX, He MN, Zhang YJ, et al. Integrating regulatory features data for prediction of functional disease associated SNPs. Brief Bioinform. 2019 Jan;20(1):26–32.
Hochberg I, Daoud D, Shehadeh N, Yom-Tov E. Can internet search engine queries be used to diagnose diabetes? Analysis of archival search data. Acta Diabetol. 2019 Oct 1;56(10):1149–54.
Hao Y, Cheng F, Pham M, Rein H, Patel D, Fang Y, et al. A noninvasive, economical, and instant-result method to diagnose and monitor type 2 diabetes using pulse wave: Case-control study. JMIR mHealth uHealth [Internet]. Shanghai Key Laboratory of Health Identification and Assessment/Laboratory of Traditional Chinese Medicine Four Diagnostic Information, Shanghai University of Traditional Chinese Medicine, Shanghai, China; 2019;7. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85066427126&doi=10.2196%2F11959&partnerID=40&md5=fa975122201fcf93db46e37135e623b9. Accessed 19 July 2020.
Reddy V, Allugunti, Elango NM, Kishor Kumar Reddy C. Internet of things based early detection of diabetes using machine learning algorithms: Dpa. Int J Innov Technol Explor Eng [Internet]. VIT University, Vellore, Tamil Nadu, India; 2019;8:1443–7. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85071304698&doi=10.35940%2Fijitee.A1013.0881019&partnerID=40&md5=dde75d7c8e703fc722145da0cdad7744.
Carter JA, Long CS, Smith BR, Smith TL, Donati GL. Combining elemental analysis of toenails and machine learning techniques as a non-invasive diagnostic tool for the robust classification of type-2 diabetes. Expert Syst Appl. 2019;115:245–55 The Boulevard, Langford Lane, Kidlington, Oxford OX5 1GB, England: Pergamon-Elsevier Science LTD;. Accessed 19 July 2020.
Seixas AA, Henclewood DA, Langford AT, McFarlane SI, Zizi F, Jean-Louis G. Differential and combined effects of physical activity profiles and Prohealth behaviors on diabetes prevalence among blacks and whites in the US population: a novel Bayesian belief network machine learning analysis. J Diabetes Res. 2017;2017:5906034. Accessed 19 July 2020.
Rodríguez-Rodríguez I, Rodríguez J-V, González-Vidal A, Zamora M-A. Feature selection for blood glucose level prediction in type 1 diabetes mellitus by using the sequential input selection algorithm (SISAL). Symmetry (Basel) [Internet]. Facultad de Informática, Departamento de Ingeniería de la Información y las Comunicaciones, Universidad de Murcia, Murcia, 30100, Spain; 2019;11. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85079617723&doi=10.3390%2FSYM11091164&partnerID=40&md5=55228bf5dbe80dd8993ccda8a0c1f618.
Baum A, Scarpa J, Bruzelius E, Tamler R, Basu S, Faghmous J. Targeting weight loss interventions to reduce cardiovascular complications of type 2 diabetes: a machine learning-based post-hoc analysis of heterogeneous treatment effects in the Look AHEAD trial. Lancet Diabetes Endocrinol England. 2017;5:808–15.
Seo W, Lee Y-B, Lee S, Jin S-M, Park S-M. A machine-learning approach to predict postprandial hypoglycemia. BMC Med Inform Decis Mak [Internet]. Department of Creative IT Engineering, POSTECH, 77, Cheongam-Ro, Nam-Gu, Pohang, 37673, South Korea; 2019;19. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85074621824&doi=10.1186%2Fs12911-019-0943-4&partnerID=40&md5=4d60d62c93df33a5a77345495c441e28. Accessed 19 July 2020.
Mohebbi A, Aradottir TB, Johansen AR, Bengtsson H, Fraccaro M, Morup M. A deep learning approach to adherence detection for type 2 diabetics. Conf Proc. Annu Int Conf IEEE Eng Med Biol Soc IEEE Eng Med Biol Soc Annu Conf [Internet]. 2017;2017:2896–9.
Rigla M, García-Sáez G, Pons B, Hernando ME. Artificial intelligence methodologies and their application to diabetes. J Diabetes Sci Technol [Internet]. Endocrinology and Nutrition Department, Parc Tauli University Hospital, Sabadell, Spain; 2018;12:303–10. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85035021972&doi=10.1177%2F1932296817710475&partnerID=40&md5=9bcbe4faecb9160fc0d31549cba108e9. Accessed 19 July 2020.
Pappada SM, Owais MH, Cameron BD, Jaume JC, Mavarez-Martinez A, Tripathi RS, et al. An artificial neural network-based predictive model to support optimization of inpatient glycemic control. Diabetes Technol Ther [Internet]. Department of Anesthesiology, University of Toledo, College of Medicine and Life Sciences, Toledo, OH 43614, United States; 2020;22:383–94. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85084595388&doi=10.1089%2Fdia.2019.0252&partnerID=40&md5=5b69e85a7d071010230d0d5f23092f9f.
Guemes A, Cappon G, Hernandez B, Reddy M, Oliver N, Georgiou P, et al. Predicting quality of overnight glycaemic control in type 1 diabetes using binary classifiers. IEEE J Biomed Heal Informatics [Internet]. Centre for Bio-inspired Technology, Imperial College London, London, United Kingdom; 2020;24:1439–46. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85084721421&doi=10.1109%2FJBHI.2019.2938305&partnerID=40&md5=197c38bc9924ead3a201cd574b11c6ea. Accessed 19 July 2020.
Acciaroli G, Sparacino G, Hakaste L, Facchinetti A, Di Nunzio GM, Palombit A, et al. Diabetes and prediabetes classification using glycemic variability indices from continuous glucose monitoring data. J Diabetes Sci Technol [Internet]. Department of Information Engineering, University of Padova, Padova, Italy; 2018;12:105–13. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85040461782&doi=10.1177%2F1932296817710478&partnerID=40&md5=2cebf7fea519e4049f75b1d2d79e5d3d. Accessed 19 July 2020.
Aliberti A, Pupillo I, Terna S, MacIi E, Di Cataldo S, Patti E, et al. A multi-patient data-driven approach to blood glucose prediction. IEEE Access [Internet]. Department of Control and Computer Engineering, Politecnico di Torino, Turin, 10129, Italy; 2019;7:69311–25. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85067264591&doi=10.1109%2FACCESS.2019.2919184&partnerID=40&md5=75e69fc728fb5a3851d8a5ff153667aa. Accessed 19 July 2020.
Hidalgo JI, Colmenar JM, Kronberger G, Winkler SM, Garnica O, Lanchares J. Data based prediction of blood glucose concentrations using evolutionary methods. J Med Syst [Internet]. Adaptive and Bioinspired System Group, School of Informatics, Universidad Complutense de Madrid, C/Profesor José García Santesmases 9, Madrid, 28040, Spain; 2017;41. Available from: https://www.scopus.com/inward/record.uri?eid=2-s2.0-85027006592&doi=10.1007%2Fs10916-017-0788-2&partnerID=40&md5=87e31929912f141ebb16b38b5526b81d. Accessed 19 July 2020.
Yang B, Wright A. Development of deep learning algorithms to categorize free-text notes pertaining to diabetes: convolution neural networks achieve higher accuracy than support vector machines. 2018; Available from: http://arxiv.org/abs/1809.05814. Accessed 19 July 2020.
Ting DSW, Cheung CY-L, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–23 Large-scale study that demonstrates deep learning systems can achieve accuracy comparable to single-human reviewers for diabetes related eye disease. Strengths include validation in large multi-ethnic cohorts. Accessed 19 July 2020.
Sosale B, Aravind SR, Murthy H, Narayana S, Sharma U, Gowda SGV, et al. Simple, Mobile-based artificial intelligence algorithm in the detection of diabetic retinopathy (SMART) study. BMJ Open Diabetes Res Care. 2020;8. Accessed 19 July 2020.
Nielsen KB, Lautrup ML, Andersen JKH, Savarimuthu TR, Grauslund J. Deep learning–based algorithms in screening of diabetic retinopathy: a systematic review of diagnostic performance. Ophthalmol Retin. 2019.
Mateen M, Wen J, Hassan M, Nasrullah N, Sun S, Hayat S. Automatic detection of diabetic retinopathy: a review on datasets, methods and evaluation metrics. IEEE Access. 2020.
Krause J, Gulshan V, Rahimy E, Karth P, Widner K, Corrado GS, et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology. 2018 Aug 1;125(8):1264–72.
Boucher MC, Qian J, Brent MH, Wong DT, Sheidow T, Duval R, et al. Evidence-based Canadian guidelines for tele-retina screening for diabetic retinopathy: recommendations from the Canadian retina research network (CR2N) tele-retina steering committee. Can J Ophthalmol. 2020 Feb 1;55(1):14–24.
Xie Y, Nguyen QD, Hamzah H, Lim G, Bellemo V, Gunasekeran DV, et al. Artificial intelligence for teleophthalmology-based diabetic retinopathy screening in a national programme: an economic analysis modelling study. The Lancet Digital Health. 2020 Apr;23.
Chandakkar PS, Venkatesan R, Li B. MIRank-KNN: multiple-instance retrieval of clinically relevant diabetic retinopathy images. J Med Imaging 2017, 4.
Aslam TM, Hoyle DC, Puri V, Bento G. Differentiation of diabetic status using statistical and machine learning techniques on optical coherence tomography angiography images. Transl Vis Sci Technol. 2020.
Cao P, Ren F, Wan C, Yang J, Zaiane O. Efficient multi-kernel multi-instance learning using weakly supervised and imbalanced data for diabetic retinopathy diagnosis. Comput Med Imaging Graph. 2018 Nov 1;69:112–24.
Saleh E, Błaszczyński J, Moreno A, Valls A, Romero-Aroca P, de la Riva-Fernández S, et al. Learning ensemble classifiers for diabetic retinopathy assessment. Artif Intell Med. 2018 Apr 1;85:50–63.
Orlando JI, Prokofyeva E, del Fresno M, Blaschko MB. An ensemble deep learning based approach for red lesion detection in fundus images. Comput Methods Prog Biomed. 2018 Jan 1;153:115–27.
Goyal M, Reeves ND, Davison AK, Rajbhandari S, Spragg J, Dfunet YMH. Convolutional neural networks for diabetic foot ulcer classification. IEEE Transactions on Emerging Topics in Computational Intelligence. 2018 Sep 12.
Zhao X, Liu Z, Agu E, Wagh A, Jain S, Lindsay C, et al. Fine-grained diabetic wound depth and granulation tissue amount assessment using bilinear convolutional neural network. IEEE Access. 2019 Dec 12;7:179151–62.
Ribeiro MT, Singh S, Guestrin C. "why should I trust you?" explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016 Aug;13:1135–44.
Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science (80-). 2019. Accessed 19 July 2020.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Diabetes Epidemiology
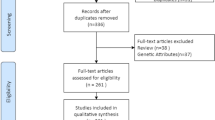
Appendix 1
Appendix 1
Article Database Search Strategies
Arxiv.org (47 total results)
-
•“Diabetes” AND “Machine Learning” in abstract, 2017–2020.
-
“Diabetes” AND “Machine Learning” in title, 2017–2020.
-
“Diabetes” AND “Random Forest*” in abstract, 2017–2020.
-
“Diabetes” AND “Decision tree*” in abstract, 2017–2020.
-
“Diabetes” AND “Support vector machine*” in abstract, 2017–2020.
-
“Diabetes” AND “Naive Bayes*” in abstract, 2017–2020.
-
“Diabetes” AND “Naïve Bayes*” in abstract, 2017–2020.
-
“Diabetes” AND “K-means clustering*” in abstract, 2017–2020.
-
“Diabetes” AND “K-nearest neighbor*” in abstract, 2017–2020.
-
“Diabetes” AND “regression tree*” in abstract, 2017–2020.
-
“Diabetes” AND “Artificial neural network*” in abstract, 2017–2020.
PubMed (70 total results).
(Search: (“Machine Learning”[tiab] OR “neural network”[tiab] OR “Deep Learning”[tiab] OR “Computer-aided”[tiab] OR “Prediction Model”[tiab] OR “Data Mining”[tiab] OR “Cross-validation” [tiab] OR “Cross validation”[tiab] OR “Regularized logistic”[tiab] OR “Linear discriminant analysis”[tiab] OR “LDA”[tiab] OR “Random forest”[tiab] OR “Naïve Bayes” [tiab] OR “Least Absolute selection shrinkage operator”[tiab] OR “elastic net”[tiab] OR “LASSO”[tiab] OR “RVM”[tiab] OR “relevance vector machine”[tiab] OR “pattern recognition”[tiab] OR “Computational Intelligence”[tiab] OR “Computational Intelligences” [tiab] OR “Machine Intelligence”[tiab] OR “Knowledge Representation”[tiab] OR “Knowledge Representations”[tiab] OR “support vector”[tiab] OR “SVM”[tiab] OR “Pattern classification” [tiab] OR “Decision Tree”[tiab] OR “Nearest neighbor”[tiab] OR “Classification Tree”[tiab] OR “Regression Tree”[tiab] OR “Iterative Learning”[tiab] OR “recursive partitioning”[tiab] OR “causal forest”[tiab] OR “generalized random forest”[tiab] OR “X-learner”[tiab] OR “R-learner”[tiab] OR “S-learner”[tiab] OR “Neural Networks, Computer”[Mesh] OR “Data Mining”[Mesh] OR “Deep Learning”[Mesh] OR “Pattern Recognition, Automated”[Mesh] OR “Artificial Intelligence”[Mesh] OR “Computational Intelligence”[Mesh] OR “Intelligence, Computational”[Mesh] OR “Machine Intelligence”[Mesh] OR “Intelligence, Machine”[Mesh] OR “Computer Reasoning”[Mesh] OR “Reasoning, Computer”[Mesh] OR “Knowledge Acquisition (Computer)”[Mesh] OR “Acquisition, Knowledge (Computer)”[Mesh] OR “Knowledge Representation (Computer)”[Mesh] OR “Knowledge Representations (Computer)”[Mesh] OR “Support Vector Machine”[Mesh] OR “Representation, Knowledge (Computer)”[Mesh] OR “Decision Trees”[Mesh])) AND (“diabetes mellitus”[mesh] OR “diabetes”[tiab]) Filters: English, from 2017 to 2020.
Scopus (497 total results).
(TITLE-ABS-KEY (“Machine Learning”)) AND (TITLE-ABS-KEY (“diabetes”)) AND (LIMIT-TO (PUBSTAGE, “final”)) AND (LIMIT-TO (DOCTYPE, “ar”)) AND (LIMIT-TO (SUBJAREA, “MEDI”) OR LIMIT-TO (SUBJAREA, “COMP”)) AND (LIMIT-TO (PUBYEAR, 2020) OR LIMIT-TO (PUBYEAR, 2019) OR LIMIT-TO (PUBYEAR, 2018) OR LIMIT-TO (PUBYEAR, 2017)) AND (LIMIT-TO (LANGUAGE, “English”))
Web of Science (161 total results).
TI = ((“Machine Learning” OR “neural network” OR “Deep Learning” OR “Computer-aided” OR “Prediction Model” OR “Data Mining” OR “Cross-validation” OR “Cross validation” OR “Regularized logistic” OR “Linear discriminant analysis” OR “LDA” OR “Random forest” OR “Naïve Bayes” OR “Least Absolute selection shrinkage operator” OR “elastic net” OR “LASSO” OR “RVM” OR “relevance vector machine” OR “pattern recognition” OR “Computational Intelligence” OR “Computational Intelligences” OR “Machine Intelligence” OR “Knowledge Representation” OR “Knowledge Representations” OR “support vector” OR “SVM” OR “Pattern classification” OR “Decision Tree” OR “Nearest neighbor” OR “Classification Tree” OR “Regression Tree” OR “Iterative Learning” OR “recursive partitioning” OR “causal forest” OR “generalized random forest” OR “X-learner” OR “R-learner” OR “S-learner” OR “Neural Networks, Computer” OR “Data Mining” OR “Deep Learning” OR “Pattern Recognition, Automated” OR “Artificial Intelligence” OR “Computational Intelligence” OR “Intelligence, Computational” OR “Machine Intelligence” OR “Intelligence, Machine” OR “Computer Reasoning” OR “Reasoning, Computer” OR “Knowledge Acquisition (Computer)” OR “Acquisition, Knowledge (Computer)” OR “Knowledge Representation (Computer)” OR “Knowledge Representations (Computer)” OR “Support Vector Machine” OR “Representation, Knowledge (Computer)” OR “Decision Trees”) AND (Diabetes)).
Rights and permissions
About this article
Cite this article
Basu, S., Johnson, K.T. & Berkowitz, S.A. Use of Machine Learning Approaches in Clinical Epidemiological Research of Diabetes. Curr Diab Rep 20, 80 (2020). https://doi.org/10.1007/s11892-020-01353-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s11892-020-01353-5