Genome-wide identification and evolution of HECT genes in wheat
- Published
- Accepted
- Received
- Academic Editor
- Zsuzsanna Dosztányi
- Subject Areas
- Agricultural Science, Bioinformatics, Genomics, Molecular Biology, Plant Science
- Keywords
- Wheat, HECT genes, Evolution, Segmental duplication, Expression
- Copyright
- © 2020 Meng et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Genome-wide identification and evolution of HECT genes in wheat. PeerJ 8:e10457 https://doi.org/10.7717/peerj.10457
Abstract
Background
As an important class of E3 ubiquitin ligases in the ubiquitin proteasome pathway, proteins containing homologous E6-AP carboxyl terminus (HECT) domains are crucial for growth, development, metabolism, and abiotic and biotic stress responses in plants. However, little is known about HECT genes in wheat (Triticum aestivum L.), one of the most important global crops.
Methods
Using a genome-wide analysis of high-quality wheat genome sequences, we identified 25 HECT genes classified into six groups based on the phylogenetic relationship among wheat, rice, and Arabidopsis thaliana.
Results
The predicted HECT genes were distributed evenly in 17 of 21 chromosomes of the three wheat subgenomes. Twenty-one of these genes were hypothesized to be segmental duplication genes, indicating that segmental duplication was significantly associated with the expansion of the wheat HECT gene family. The Ka/Ks ratios of the segmental duplication of these genes were less than 1, suggesting purifying selection within the gene family. The expression profile analysis revealed that the 25 wheat HECT genes were differentially expressed in 15 tissues, and genes in Group II, IV, and VI (UPL8, UPL6, UPL3) were highly expressed in roots, stems, and spikes. This study contributes to further the functional analysis of the HECT gene family in wheat.
Introduction
Ubiquitination is a post-translational modification that involves the covalent attachment of ubiquitin to a protein substrate. Ubiquitination is important for cellular homeostatic maintenance and plays essential roles in plant growth, development, and the regulation of abiotic and biotic stresses (Downes et al., 2003; El Refy et al., 2003; Liu & Stone, 2011; Miao & Zentgraf, 2010; Moon, Parry & Estelle, 2004; Rotin & Kumar, 2009; Stone, 2014; Wang & Deng, 2011). At the end of a three-enzyme cascade (E1 ubiquitin activating enzyme; E2 ubiquitin conjugating enzyme; E3 ubiquitin ligase), E3 recruits substrates and promotes or directly catalyzes ubiquitin transfer onto its targets (Huibregtse et al., 1995; Scheffner, Nuber & Huibregtse, 1995; Wang et al., 2006; Wang & Pickart, 2005). E3 generally determines the specificity of the ubiquitination reaction with different substrate recognition domains (Huibregtse et al., 1995; Kim et al., 2007; Scheffner, Nuber & Huibregtse, 1995). According to structural similarities and ubiquitination domains, plant E3 proteins can be classified as three main types (Chen & Hellmann, 2013; Craig et al., 2009; Duplan & Rivas, 2014; Guzman, 2014; Huibregtse et al., 1995; Mach, 2008; Maspero et al., 2013; Qin et al., 2008; Scheffner, Nuber & Huibregtse, 1995; Schwechheimer & Calderon Villalobos, 2004; Stone, 2014; Wang et al., 2006; Wang & Pickart, 2005; Yee & Goring, 2009): HECT (Homologous to E6-AP C-Terminal), RING (Really Interesting New Gene), and U-box.
The HECT-type ubiquitin ligase is an important class of E3s defined by the presence of a C-terminal catalytic HECT domain. The general features of HECT domains are an N-terminal lobe that contains the E2-binding site and a smaller C-terminal lobe that includes the active-site Cys residue that receives ubiquitin from E2 and links itself with the ubiquitin molecule (Downes et al., 2003; Huibregtse et al., 1995). Classification of HECT E3 proteins into different subfamilies is based on the N-terminal domains (Downes et al., 2003; Grau-Bove, Sebe-Pedros & Ruiz-Trillo, 2013; Marin, 2010; Marin, 2013) responsible for recognizing and binding protein substrates (Kamadurai et al., 2013; Kim et al., 2011; Maspero et al., 2011; Maspero et al., 2013; Rotin & Kumar, 2009), while the conserved C-terminal HECT domain catalyzes the transfer of ubiquitin to various substrates. Substrate proteins usually possess recognition motifs that can directly bind to the N-terminal domains, while the special HECT domains are essential to the prediction and evolution of the HECT genes in plants; however, comprehensive research on these genes is limited.
The HECT-type E3 ubiquitin ligases comprise a small class of E3s, and seven genes (UPL1-UPL7) have been identified in Arabidopsis thaliana (Downes et al., 2003). UPL3 is involved in trichome development (Downes et al., 2003; Patra, Pattanaik & Yuan, 2013), genome endoreduplication (El Refy et al., 2003) and seed size (Miller et al., 2019). UPL5 is involved in leaf senescence (Miao & Zentgraf, 2010), and UPL1, UPL3, and UPL5 in plant immunity (Furniss et al., 2018). These seven A. thaliana HECT genes can be classified into five subfamilies or six groups according to the phylogenetic relationships provided in previous studies (Marin, 2013; Meng et al., 2015). However, little research has been conducted on the HECT genes in wheat, which is one of the most important crops produced worldwide (Choulet et al., 2014; International Wheat Genome Sequencing Consortium, 2014; International Wheat Genome Sequencing Consortium et al., 2018). In this research, we conducted a comprehensive genome-wide analysis of the wheat HECT genes to identify HECT genes conserved in wheat, rice, and A. thaliana. Gene exon-intron structure, conserved motif, domain structure, chromosomal distribution, duplication event, and expression profile were also analyzed in detail. Our research data will provide useful information for further functional investigation of the HECT gene family in allohexaploid wheat and their evolution in polyploid plants.
Materials & Methods
Sequence retrieval and identification of the HECT gene family in wheat
To identify the HECT genes in wheat, the protein sequences of all HECT genes in A. thaliana and rice were retrieved from the Phytozome v13 database (Goodstein et al., 2012) with the Ensembl Plants (Howe et al., 2020) as a complementary sequence database. These protein sequences were then used as queries to conducted local BlastP and tBlastN (Camacho et al., 2009) searches using default parameters (E-value <10−5) against wheat reference sequence database in the Chinese Spring reference IWGSC RefSeq v1.0 (International Wheat Genome Sequencing et al. 2018) from the Ensembl Plants database. The hmmsearch program in the HMMER software (version 3.3) (Potter et al., 2018) was also used to the identification of HECT genes with the HMM profile of the HECT domain (PF00632) in the Pfam 32.0 database (El-Gebali et al., 2019), using the default parameters (E-value < 10−5). Then, the combined candidate HECT genes were used as queries to conduct BlastP and tBlastN searches of the wheat genome again to obtain more potential gene candidates with the default parameters (E-value < 10−5). The obtained protein sequences were further verified using the InterProScan program (Jones et al., 2014) to confirm the presence of the HECT domain. Finally, each HECT gene was revised manually for conserved domain architectures using the Pfam (El-Gebali et al., 2019), PROSITE (Sigrist et al., 2013), and SMART (Letunic & Bork, 2018) databases. Proteins without a typical HECT domain or fewer than 300 amino acids were removed from the final sequence dataset.
Sequence alignment and phylogenetic analysis
Multiple sequence alignments of the wheat HECT protein sequences were performed by using MUSCLE (Edgar, 2004) with its default parameters, and MAFFT (L-INS-i strategy) (Rozewicki et al., 2019). The phylogenetic tree was constructed and visualized using MEGAX software (Kumar et al., 2018) based on the full-length HECT protein sequences through a neighbor-joining algorithm with 1,000 bootstrap repetitions. The maximum likelihood (ML) methods implemented in PhyML3.1 (Guindon et al., 2010) were also used to construct trees of full-length HECT protein sequences with 1,000 bootstrap repetitions.
Sequence analysis
The structures of HECT genes and the number of exons and introns were determined using the Gene Structure Display Server (Hu et al., 2015) by aligning the coding sequences with their corresponding genomic DNA sequences. The conserved motifs encoded by HECT genes were identified using MEME (Multiple EM for Motif Elicitation) (Bailey et al., 2015). The conserved domains of the HECT protein sequences within the phylogenetic trees were visualized and annotated using EvolView (Subramanian et al., 2019).
Chromosomal location and duplication
To map all HECT genes to the wheat chromosomes, information of HECT gene chromosomal location was obtained from Ensembl Plants. Gene duplication events of the wheat HECT genes were inferred based on their location among the three wheat subgenomes (A, B, and D). Firstly, all-in-all BlastP of the wheat genome was performed to analyze sequence similarity among the three subgenomes (A, B, and D). Secondly, MCScanX (Multiple Collinearity Scan toolkit) (Wang et al., 2012) was then used with default parameters to detect possible gene duplication blocks. Finally, Chromosomal location and syntenic relationships were illustrated using Circos-0.67 (Krzywinski et al., 2009). Synonymous (Ks) and nonsynonymous substitution (Ka) rates were calculated with TBtools (Chen et al., 2018), as previously described (Meng et al., 2015). For each gene pair, the approximate divergence time (T, million years ago, Mya) of the duplication events for each paralogous gene pair was estimated using the mean Ks values from T = Ks/2 λ, in which the mean synonymous substitution rate (λ) for wheat is 6 ×10−9(Wolfe, Li & Sharp, 1987; Wolfe, Sharp & Li, 1989).
Expression analyses
RNA-Seq data (Accession number “choulet_URGI” and “PRJNA497810”) were downloaded from expVIP (Borrill et al., 2019; Borrill, Ramirez-Gonzalez & Uauy, 2016; Choulet et al., 2014; Ramírez-González et al., 2018) (Table S1–S4) and used to explore the expression patterns of HECT genes in wheat. These transcript data were obtained from five organs (root, stem, leaf, spike, and grain) at three developmental stages and flag leaves harvested at 3, 7, 10, 13, 15, 17, 19, 21, 23, and 26 dpa (day post-anthesis). The expression data were log2 based TPM (transcripts per million mapped reads) values, and the heatmap of the expression patterns of wheat HECT genes was drawn using the R heatmap.2 function.
Results
Identification of the HECT gene family in wheat
To identify HECT genes in wheat, the HMM HECT domain profile PF00632 (El-Gebali et al., 2019) and the HECT protein sequences from A. thaliana (Downes et al., 2003) and rice (Meng et al., 2015) were used to search against the wheat protein sequences in Ensemble Plants (Howe et al., 2020; International Wheat Genome Sequencing et al. 2018), then the potential HECT genes were confirmed by InterProScan (Jones et al., 2014) with Pfam (El-Gebali et al., 2019), Prosite (Sigrist et al., 2013) and SMART (Letunic & Bork, 2018) databases that helped to characterize the candidates by the existence of the complete HECT domain. Ultimately, we identified 25 putative HECT genes in the latest wheat genome (Table 1).
| Gene symbol | Gene locus | Chromosome | Gene start (bp) | Gene end (bp) | Gene % GC content | length (AA) |
|---|---|---|---|---|---|---|
| TaHECT01 | TraesCS1A02G059500 | 1A | 40892389 | 40898032 | 47.13 | 1335 |
| TaHECT02 | TraesCS1A02G106100 | 1A | 103311704 | 103326325 | 42.01 | 1010 |
| TaHECT03 | TraesCS1A02G288600 | 1A | 485557978 | 485566303 | 43.98 | 1520 |
| TaHECT04 | TraesCS1B02G123400 | 1B | 149601252 | 149613588 | 39.18 | 1027 |
| TaHECT05 | TraesCS1B02G298000 | 1B | 518258672 | 518266604 | 42.38 | 1521 |
| TaHECT06 | TraesCS1D02G108900 | 1D | 101929905 | 101943199 | 39.16 | 1030 |
| TaHECT07 | TraesCS1D02G287600 | 1D | 386119575 | 386127952 | 44.18 | 1522 |
| TaHECT08 | TraesCS2A02G064700 | 2A | 29095129 | 29105808 | 42.87 | 1862 |
| TaHECT09 | TraesCS2B02G076900 | 2B | 42143087 | 42153426 | 43.11 | 1835 |
| TaHECT10 | TraesCS3B02G194900 | 3B | 215607542 | 215618805 | 42.3 | 695 |
| TaHECT11 | TraesCS4A02G285800 | 4A | 591699693 | 591709939 | 41.24 | 1036 |
| TaHECT12 | TraesCS4B02G027200 | 4B | 20589014 | 20599094 | 41.07 | 1170 |
| TaHECT13 | TraesCS4D02G025000 | 4D | 10931453 | 10942048 | 41.32 | 1122 |
| TaHECT14 | TraesCS5A02G121600 | 5A | 263498258 | 263513761 | 42.56 | 3632 |
| TaHECT15 | TraesCS5A02G262600 | 5A | 475656292 | 475662357 | 45.58 | 832 |
| TaHECT16 | TraesCS5B02G112800 | 5B | 177183624 | 177199346 | 42.35 | 3628 |
| TaHECT17 | TraesCS5B02G261000 | 5B | 445453118 | 445459346 | 44.89 | 858 |
| TaHECT18 | TraesCS5D02G118000 | 5D | 154524415 | 154539554 | 42.35 | 3631 |
| TaHECT19 | TraesCS5D02G270200 | 5D | 373832243 | 373838247 | 44.01 | 817 |
| TaHECT20 | TraesCS6A02G003300 | 6A | 1494577 | 1504714 | 46.25 | 1839 |
| TaHECT21 | TraesCS6B02G000300 | 6B | 163680 | 173488 | 45.87 | 1851 |
| TaHECT22 | TraesCS6D02G005600 | 6D | 2399592 | 2409297 | 45.55 | 1841 |
| TaHECT23 | TraesCS7A02G244000 | 7A | 220125320 | 220130919 | 46.62 | 862 |
| TaHECT24 | TraesCS7B02G313300 | 7B | 559527374 | 559559604 | 45.86 | 897 |
| TaHECT25 | TraesCS7B02G496200 | 7B | 747214104 | 747221564 | 42.45 | 796 |
Phylogenetic analysis of HECT genes in wheat
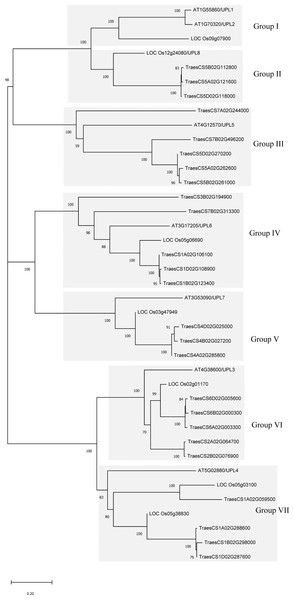
To understand the evolutionary relationship of the wheat HECT genes, phylogenetic trees were constructed based on the alignment of the full-length protein sequences and HECT domain sequences of 25 wheat, 7 rice, and 7 A. thaliana HECT proteins (Fig. 1 and Fig. S1). According to the classification criteria used for A. thaliana and rice in previous studies (Downes et al., 2003; Grau-Bove, Sebe-Pedros & Ruiz-Trillo, 2013; Marin, 2013; Meng et al., 2015), the wheat HECT genes were categorized into seven groups (Group I, II, III, IV, V, VI and VII), which contained 0, 3, 5, 5, 3, 5 and 4 HECT genes, respectively. Genes in Group III, IV and VI were the most abundant and comprised 60% of the identified genes, while genes in Group I was absent in wheat. Nevertheless, in A. thaliana, Group I included two HECT genes, Group II did not include any HECT genes, and other Groups included one HECT gene, respectively. These seven groups can be further classified into five subfamilies that correspond to those described in a previous study (Marin, 2013).
Figure 1: Phylogenetic relationships of 39 HECT genes from wheat (25), rice (seven), and Arabidopsis thaliana (seven).
A neighbor-joining (NJ) unrooted tree is shown and the shaded areas indicate the main branches that correspond to the seven gene groups. MEGAX package was used to construct the NJ tree from full-length amino acid sequence alignments (File S1) of the three plant species, with 1000 bootstrap replicates. Numbers refer to bootstrap support in terms of percentage.Gene exon-intron structure and conserved motif and domain architecture of the wheat HECT genes
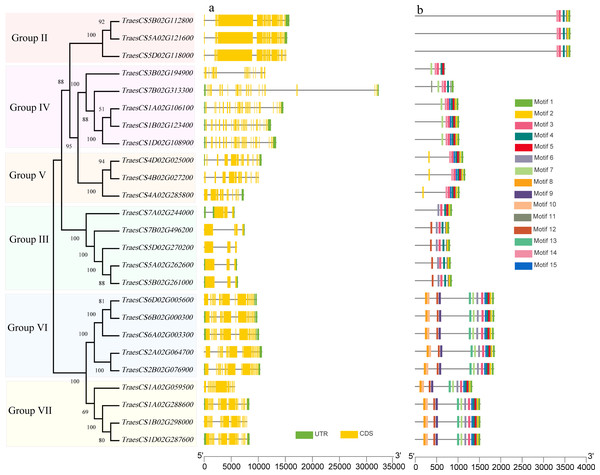
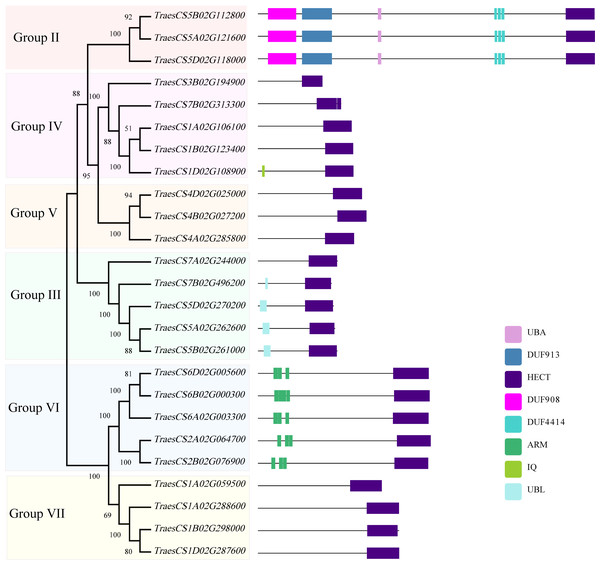
To investigate the structural characteristics of wheat HECT genes, the exon-intron structures of the wheat HECT genomic sequences, conserved motifs, and the domain architecture of the wheat HECT proteins were compared based on their phylogenetic relationships. Each gene structure was revealed by aligning its coding sequences with the corresponding genomic sequences (Chen et al., 2018; Hu et al., 2015). Most of the wheat HECT genes contained abundant (more than ten) exons and only those in Group III had only three or four exons (Fig. 2A). Closely related HECT genes in the same phylogenetic group had similar exon-intron structures, and those with closer evolutionary relationships were more similar in their number and length of exons and introns. The conserved motifs of wheat HECT proteins in each group were analyzed using MEME software (Bailey et al., 2015). Fifteen conserved motifs (motif1-motif15) were predicted and these motifs were specific to each group (Fig. 2B). The composition of the structural motifs varied among the different HECT groups, while similar motifs were found in the same group. Additionally, the motifs encoding the HECT domain in the C-terminal regions of wheat HECT proteins were relatively conserved, suggesting that the functions of the HECT proteins were intergroup specific. The domain architecture of HECT proteins was obtained using the InterProScan program (Jones et al., 2014) with a three-database annotation (El-Gebali et al., 2019; Letunic & Bork, 2018; Sigrist et al., 2013). In addition to the HECT domain, other domains were found in the N-terminal regions of wheat HECT proteins (Fig. 3). The wheat HECT genes that were derived from the same group generally had similar exon-intron structures (Fig. 2A), motif compositions (Fig. 2B), and domain architectures (Fig. 3).
Figure 2: Gene structures and conserved motifs that encode 25 wheat HECT proteins based on phylogenetic relationships.
The unrooted neighbor-joining tree was constructed using the alignment of full-length amino acid sequences (File S2) with the MEGAX package. The lengths of the exons and introns in each HECT gene are displayed proportionally. The green boxes, yellow boxes, and lines indicate UTRs, exons, and introns, respectively. Conserved motifs are showed in different colors.Figure 3: Domain architecture of 25 wheat HECT proteins according to their phylogenetic relationships.
Each domain is represented by a colored box. UBA: Ubiquitin associated domain, DUF: Domain of Unknown Function, ARM: Armadillo repeats, IQ; IQ Short calmodulin-binding motif, UBL: Ubiquitin-like domain.Chromosomal location and duplication of wheat HECT genes
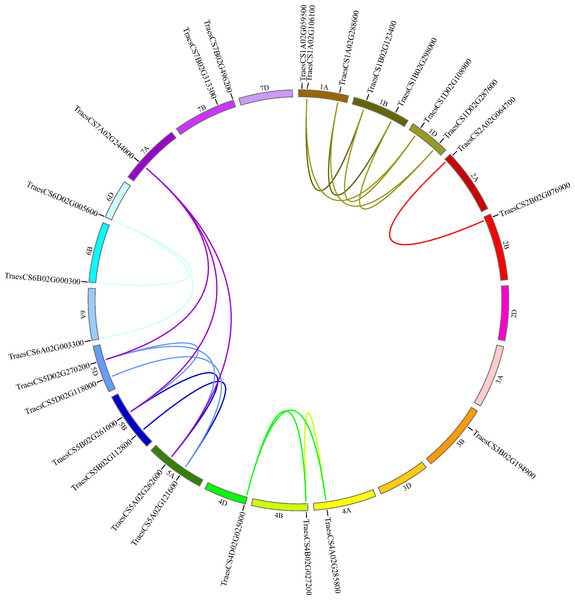
To decide the chromosomal locations of the wheat HECT genes, the 25 putative wheat HECT genes were located in the 21 chromosomes of the wheat genome database available from Ensembl Plants (Howe et al., 2020; International Wheat Genome Sequencing et al. 2018). The wheat HECT genes were randomly distributed in 17 of 21 chromosomes; chromosome 2D, 3A, 3D, and 7D contained no HECT genes, chromosome 1A contained three HECT genes, chromosome 1B, 1D, 5A, 5B, 5D, and 7B each contained two HECT genes, and the other chromosomes each contained only one HECT gene (Fig. 4). The 25 wheat HECT genes were approximately evenly distributed among the A (9), B (10), and D (6) subgenomes, which was in accordance with the observation that most HECT genes have three homoeologous sequences located across three subgenomes. However, the HECT genes were not randomly distributed among the different chromosomal groups of the three subgenomes. The chromosomal Group II, III, and VII contained two, one, and three sequences, respectively. The remaining 19 sequences were more evenly distributed across chromosomal Group I, IV, V, and VI, and ranged from three to seven genes per group. An interesting finding was that the location of the HECT genes on chromosome 4A was opposite with those of the homoeologous genes on chromosome 4B and chromosome 4D (Fig. 4).
Figure 4: Chromosome locations of HECT genes and segmentally duplicated gene pairs in the wheat genome.
Chromosomes are shown in different colors and in a circular form. The approximate distribution of each wheat HECT gene is marked on the circle with a short black line. Colored curves denote the details of syntenic regions between the wheat HECT genes. The purple curves represent the estimated time of duplication events that occurred 100–112 Mya, and the other curves represent the estimated time of duplication events 2–12 Mya.Segmental and tandem duplication are considered two essential factors for gene family expansion in plants (Cannon et al., 2004; Panchy, Lehti-Shiu & Shiu, 2016; Qiao et al., 2019; Zhu et al., 2014). To examine duplication patterns of the wheat HECT genes, we identified tandem and segmental duplication events using MCscanX (Wang et al., 2012) employing default parameters with TBtools (Chen et al., 2018). No tandem duplicated HECT gene pairs were identified in the 25 wheat HECT genes; however, 21 of the 25 wheat HECT genes were involved in segmental duplication. Twenty segmental duplicated HECT gene pairs were identified (Fig. 4 and Table 2), indicating that the segmental duplication events had contributed to HECT gene family expansion. To date the gene duplication time of these segmentally duplicated HECT genes, the Ks and Ka distances, as well as the Ka/Ks ratios were calculated. The Ka/Ks ratios for segmentally duplicated HECT gene pairs ranged from 0.07 to 0.44, with an average value of 0.20 (Table 2), implying that these segmentally duplicated HECT genes were under purifying selection, as indicated by the Ka/Ks ratios were less than 1. The divergence time of duplication events were inferred by Ks (Table 2). Results showed that within six existed phylogenetic groups, the two closest wheat HECT genes were duplicated about 2–12 million years ago (Mya), while the other genes were duplicated about 100–112 Mya.
| Group | Gene 1 | Gene 2 | Ka | Ks | Ka/Ks | Mya |
|---|---|---|---|---|---|---|
| II | TraesCS5A02G121600 | TraesCS5B02G112800 | 0.01 | 0.04 | 0.16 | 2.93 |
| TraesCS5A02G121600 | TraesCS5D02G118000 | 0.00 | 0.03 | 0.14 | 2.87 | |
| III | TraesCS5A02G262600 | TraesCS5B02G261000 | 0.01 | 0.08 | 0.18 | 6.29 |
| TraesCS5A02G262600 | TraesCS5D02G270200 | 0.01 | 0.07 | 0.18 | 6.09 | |
| TraesCS5A02G262600 | TraesCS7A02G244000 | 0.55 | 1.34 | 0.41 | 111.99 | |
| TraesCS5B02G261000 | TraesCS5D02G270200 | 0.01 | 0.06 | 0.23 | 4.79 | |
| TraesCS5B02G261000 | TraesCS7A02G244000 | 0.55 | 1.29 | 0.42 | 107.74 | |
| TraesCS5D02G270200 | TraesCS7A02G244000 | 0.55 | 1.24 | 0.44 | 103.15 | |
| IV | TraesCS1A02G106100 | TraesCS1B02G123400 | 0.00 | 0.04 | 0.07 | 3.07 |
| TraesCS1A02G106100 | TraesCS1D02G108900 | 0.00 | 0.03 | 0.15 | 2.44 | |
| TraesCS1B02G123400 | TraesCS1D02G108900 | 0.00 | 0.03 | 0.17 | 2.29 | |
| V | TraesCS4A02G285800 | TraesCS4B02G027200 | 0.01 | 0.03 | 0.19 | 2.91 |
| TraesCS4A02G285800 | TraesCS4D02G025000 | 0.02 | 0.05 | 0.30 | 4.37 | |
| TraesCS4B02G027200 | TraesCS4D02G025000 | 0.01 | 0.06 | 0.24 | 5.04 | |
| VI | TraesCS2A02G064700 | TraesCS2B02G076900 | 0.00 | 0.03 | 0.15 | 2.66 |
| TraesCS6A02G003300 | TraesCS6D02G005600 | 0.01 | 0.12 | 0.08 | 10.01 | |
| TraesCS6B02G000300 | TraesCS6D02G005600 | 0.01 | 0.13 | 0.07 | 11.03 | |
| VII | TraesCS1A02G288600 | TraesCS1B02G298000 | 0.01 | 0.06 | 0.23 | 5.41 |
| TraesCS1A02G288600 | TraesCS1D02G287600 | 0.01 | 0.05 | 0.11 | 4.25 | |
| TraesCS1B02G298000 | TraesCS1D02G287600 | 0.01 | 0.08 | 0.17 | 6.43 |
Notes:
- Ks
-
synonymous substitution rate
- Ka
-
nonsynonymous substitution rate
- Mya
-
million years ago
Expression profiles of wheat HECT genes
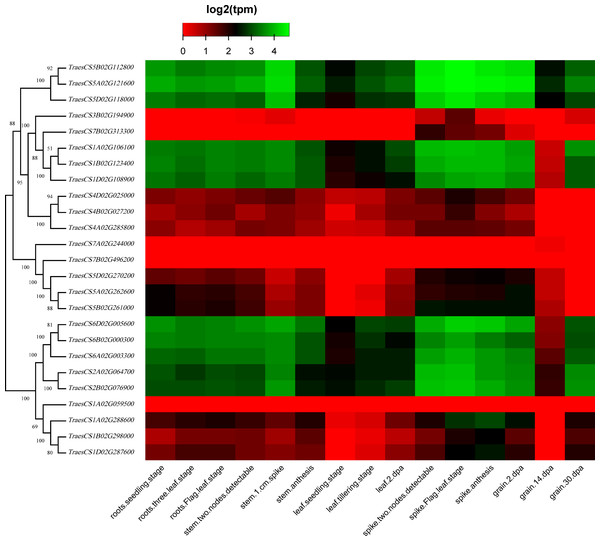
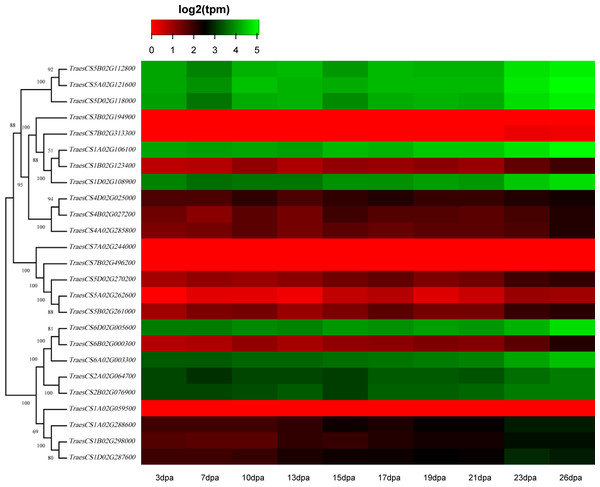
To discover the potential roles of these wheat HECT genes in growth and development, we used public RNA-seq data covering 15 tissues at different growth stages from expVIP (Borrill et al., 2019; Borrill, Ramirez-Gonzalez & Uauy, 2016; Ramírez-González et al., 2018). Based on the wheat RNA-seq data, the 25 wheat HECT genes were detected in all 15 tissues at the gene level (Fig. 5, Table S1, and Table S2). Moreover, the expression of these genes exhibits distinct expression and tissue-specific characteristics. Most HECT genes in Group II, IV, and VI were relatively highly expressed in the roots, stems and spikes, while those in the leaves were expressed at relatively low levels (Fig. 5). Interestingly, in wheat grain tissues, most wheat HECT gene expression in Group II, IV, and VI were high at 2 dpa and 30 dpa and low at 14 dpa. Moreover, genes within each group or in different groups had similar expression patterns in different tissues, such as the high expression of genes in Group II (TraesCS5A02G121600, TraesCS5B02G112800, TraesCS5D02G118000), Group IV (TraesCS1A02G106100, TraesCS1B02G123400, TraesCS1D02G108900), and Group VI (TraesCS6A02G003300, TraesCS6B02G000300, TraesCS6D02G005600, TraesCS2A02G064700, and TraesCS2B02G076900), except for TraesCS3B02G194900 and TraesCS7B02G313300 (Fig. 5). Furthermore, the genes in Group II, IV, and VI were relatively highly expressed in the spikes at different developmental stages and in stems at the one cm spike stage compared to those in other tissues (Fig. 5). According to RNA-seq data of the ten-time point expression time course of wheat senescence in the flag leaf, the expression level of most wheat HECT genes in Group II, IV, and VI gradually increased with the increase of dpa (Fig. 6, Table S3, and Table S4).
Figure 5: Heatmap of wheat HECT gene expression patterns in 15 different tissues.
Transcriptional levels were obtained from expVIP. The RNA-seq relative expression data from 15 tissues was used to reconstruct the expression patterns of the wheat genes. The sources of the samples are provided on the x-axis.Figure 6: Heatmap of wheat HECT gene expression patterns in wheat leaf senescence.
Transcriptional levels were obtained from expVIP. The RNA-seq relative expression data from flag leaves harvested at 3, 7, 10, 13, 15, 17, 19, 21, 23, and 26 dpa (day post-anthesis) was used to reconstruct the expression patterns of the wheat genes. The sources of the samples are provided on the x-axis.Discussion
HECT genes play important roles in A. thaliana and diverse plant growth, developmental and physiological processes (Downes et al., 2003; (El Refy et al., 2003); Furniss et al., 2018; Miao & Zentgraf, 2010; Miller et al., 2019; Patra, Pattanaik & Yuan, 2013), including trichome development (Downes et al., 2003), genome endoreduplication (El Refy et al., 2003), seed size (Miller et al., 2019), leaf senescence (Miao & Zentgraf, 2010), and plant immunity (Furniss et al., 2018). However, this gene family has not been investigated in wheat. In this research, we conducted an extensive analysis of the wheat HECT genes, including their evolution, gene exon-intron structure, conserved motif, domain structure, chromosomal location, duplication event, and expression pattern.
We identified 25 HECT genes in the wheat genome, which is 3.6 times the number present in A. thaliana (Downes et al., 2003). However, a former study discovered 19 soybean HECT genes, which is 2.7 times the number found in A. thaliana (Meng et al., 2015). Our results showed six more HECT genes in wheat than the number previously estimated in the soybean genome. A possible explanation for this difference is that wheat is a hexaploid crop with 21 chromosomes containing three subgenomes (A, B, and D) (International Wheat Genome Sequencing Consortium, 2014; International Wheat Genome Sequencing Consortium et al., 2018), while soybean is a diploid crop with 20 chromosomes derived from an ancient tetraploid that may have had about two times more the number of HECT genes than other normal diploid species (Schmutz et al., 2010).
The phylogenetic analysis of the 25 wheat HECT genes classified them into subfamilies similar to those characterized by previous research (Downes et al., 2003; Grau-Bove, Sebe-Pedros & Ruiz-Trillo, 2013; Marin, 2013; Meng et al., 2015). The classification was according to the corresponding HECT gene sequence homology. Based on the phylogenetic relationships among the HECT genes in wheat, rice, and A. thaliana, the wheat HECT genes were classified into seven groups. Compared with a former report in A. thaliana (Marin, 2013), subfamily IV HECT genes were absent in wheat. Wheat subfamily V (Group I, UPL1/2 and Group II, UPL8 in this study) contained three genes, subfamily VI (Group III, UPL5) contained five genes, subfamily III (Group IV, UPL6) contained five genes, Subfamily II (Group V, UPL7) contained three genes, and subfamily I (Group VI, UPL3 and Group VII, UPL4) contained nine genes. The HECT gene Group I was not observed in the wheat genome. With the exception of Group I and II, other wheat Groups own HECT genes orthologous with A. thaliana. This is basically consistent with the results of a previous HECT gene investigation in plants (Marin, 2013), suggesting that A. thaliana HECT gene Group II (UPL8 in this study) was lost, while the wheat HECT gene Group I (UPL1/UPL2) was not observed in our analysis. Gene members of each phylogenetic Group often possess identical gene exon-intron structures, conserved motifs, and domain architectures, indicating that they probably recognize, bind, and might interact with same or similar substrate protein.
Segmental duplication events, tandem duplication events, as well as transposition events are three main evolutionary mechanisms of duplication events that expand the members of gene family (Cannon et al., 2004; Panchy, Lehti-Shiu & Shiu, 2016; Qiao et al., 2019; Zhu et al., 2014). Segmental duplications frequently occur in higher plants, because they are diploidized polyploids that have maintained various duplicated chromosomal blocks in the existing genomes (Cannon et al., 2004; Qiao et al., 2019). In this present research, we discovered that 21 of the 25 wheat HECT genes were located in chromosomes across the three subgenomes (A, B, D), indicating that segmental duplication obviously contributed to expanding the wheat HECT gene family. A previous study has shown that the allohexaploid wheat subgenomes A, B, and D were originally derived from three diploid (2x; 2n = 14) species and underwent three hybridization events (International Wheat Genome Sequencing 2014). The A and B subgenomes diverged from a common ancestor ∼7 million years ago and the first hybridization occurred ∼5.5 million years ago between A and B subgenomes, leading to the D subgenome through homoploid hybrid speciation. The second hybridization between the A and B subgenomes gave rise to the AABB genome <0.8 million years ago via polyploidization. Wheat originated <0.4 million years ago by allopolyploidization from a third hybridization. By estimating the approximate dates of the segmentally duplicated pairs of wheat HECT genes, we infer that the paralogous genes in wheat HECT groups originated from a relatively recent duplication event during the shaping of the three subgenomes (A, B, D) that occurred before the second hybridization event in wheat evolution history, except for TraesCS7A02G244000 in Group III, which originated from a relatively ancient duplication event before the appearance of the common ancestor of the A and B subgenomes. Thus, segmental duplication events were the primary driving forces for HECT gene evolution during the speciation and evolution of allohexaploid wheat.
To better understand the roles of the HECT genes during the life cycle of wheat, we performed an expression analysis of public RNA-seq data (Borrill, Ramirez-Gonzalez & Uauy, 2016; Choulet et al., 2014; Ramírez-González et al., 2018) in 15 tissues at different developmental stages. Analysis of the expression patterns of these wheat genes in 15 tissues showed that most wheat HECT genes in Group II, IV, and VI were relatively highly expressed in the roots, stems, and spikes. In particular, the genes in Groups II, IV, and VI were relatively highly expressed in the spikes at different developmental stages and in stems at the 1-cm spike stage compared to other tissues. Therefore, the expression of these genes may be closely related to wheat spike growth and development, suggesting that the HECT genes in highly expressed spikes may be involved in the regulation or degradation of proteins via ubiquitination during spike development stage. Previous studies have revealed that A. thaliana AT4G38600/UPL3 plays a specific role during trichome development (Downes et al., 2003; Patra, Pattanaik & Yuan, 2013) and seed size (Miller et al., 2019) and that AT4G12570/UPL5 is an important transcription factor that positively regulates leaf senescence by the ubiquitination and degradation of AT4G23810/WRKY5 3 (Miao & Zentgraf, 2010). In our investigation, the wheat genes orthologous to A. thaliana AT4G38600/UPL3 included five paralogous genes in Group VI. Except for grain at the 14 dpa stage, these five genes were all relatively highly expressed in wheat, particularly in spikes. A reasonable explanation is that the relatively low expression of UPL3 at 14 dpa stage may be related to the size of wheat seeds and is an adaptive regulation mechanism during seed formation. This is consistent with a recent study of UPL3 in Brassica napus (Miller et al., 2019). Miller et al. determined a mechanism in which the proteasomal degradation of LEC2, a transcription factor controlling seed maturation, is mediated by UPL3 and reduced UPL3 expression would increase LEC2 protein levels and seed size. The wheat genes orthologous to A. thaliana AT4G12570/UPL5 were five paralogous genes in Group III, which were expressed in different wheat tissues but showed distinct features. At different developmental stages, the expression levels of TraesCS5D02G270200, TraesCS5A02G262600, and TraesCS5B02G261000 in roots, stems, and spikes were relatively unchanged, but gradually increased in leaves and decreased in grain. The three wheat genes TraesCS1A02G106100, TraesCS1B02G123400, and TraesCS1D02G108900 are orthologous to A. thaliana AT3G17205/UPL6 in Group IV, and the genes in Group II (UPL8 absent in A. thaliana) showed similar expression patterns to those in Group VI (UPL3). RNA-seq data of wheat leaf senescence (Borrill et al., 2019) indicated that HECT genes in Group II, IV and VI (UPL8, UPL6, and UPL3) might also play crucial roles in plant leaf senescence. The differential expression of paralogous HECT genes in or among groups in wheat suggests that they might have the same or similar functions as their orthologous genes in A. thaliana and Brassica napus; nevertheless, they might have evolved functional differences.
A former research discovered that AT4G38600/UPL3 mediated UPS-dependent proteolysis of the two transcription factors AT5G41315/GL3 and AT1G63650/EGL3, which interact with the ARM domains of UPL3 and function as positive regulators during A. thaliana trichome development (Patra, Pattanaik & Yuan, 2013). The evolutionarily closely related Group VII (UPL4) and VI (UPL3) belong to the same subfamily I defined in previous studies (Grau-Bove, Sebe-Pedros & Ruiz-Trillo, 2013; Marin, 2013) and genes in Group VII contain no ARM domains (Fig. 3) and thus, are differentially expressed at relatively low levels (Fig. 5) compared to those genes in Group VI. More functional explorations of these genes could improve our understanding of the roles of HECT genes in wheat and other plants during growth and development.
Conclusions
Herein, 25 identified wheat HECT genes were classified into six phylogenetic groups and distributed evenly in 17 of 21 chromosomes of the three subgenomes. Twenty-one hypothesized segmentally duplicated genes indicated that segmental duplication was significantly associated with the expansion of these HECT genes. The expression analysis revealed that most wheat HECT genes in Group II, IV, and VI (UPL8, UPL6, and UPL3) were highly expressed in roots, stems, and spikes at different developmental stages and gradually increased with the increase of dpa, suggesting that these genes may be involved in wheat growth, development and leaf senescence. This study provides useful information for further biological functional analysis of the HECT gene family in allohexaploid wheat.
Supplemental Information
Phylogenetic relationships of 39 HECT genes from wheat (25), rice (7), and Arabidopsis thaliana (7)
A neighbor-joining (NJ) unrooted tree is shown and the shaded areas indicate the main branches that correspond to the seven gene groups. MEGAX package was used to construct the NJ tree from domain sequence alignments (File S4) of the three plant species, with 1000 bootstrap replicates. Numbers refer to bootstrap support in terms of percentage.