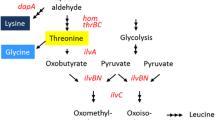
Abstract
The psefdh_D221Q gene encodes a mutant formate dehydrogenase from Pseudomonas (P-seFDG_D221Q), which catalyzes formate oxidation with the simultaneous formation of NADPH. It is expressed in cells of lysine-producing Corynebacterium glutamicum strains. The psefdh_D221Q gene was introduced into C. glutamicum strains as part of an autonomous plasmid or was integrated into the chromosome with the simultaneous inactivation of the host formate-dehydrogenase genes. It was shown that the C. glutamicum strains with NADP+-dependent formate dehydrogenase have an increased level of L-lysine synthesis in the presence of formate if their own formate dehydrogenase is inactivated.

Similar content being viewed by others
REFERENCES
Kjeldsen, K. and Nielsen, J., In silico genome-scale reconstruction and validation of the Corynebacterium glutamicum metabolic network, Biotechnol. Bioeng., 2009, vol. 102, no. 2, pp. 583–597. https://doi.org/10.1002/bit.22067
Melzer, G., Esfandabadi, M.E., Franco-Lara, E., and Wittmann, C., Flux Design: In silico design of cell factories based on correlation of pathway fluxes to desired properties, BMC Syst Biol., 2009, vol. 3, p. 120. https://doi.org/10.1186/1752-0509-3-120
Becker, J. and Wittmann, C., Systems and synthetic metabolic engineering for amino acid production—the heartbeat of industrial strain development, Curr. Opin. Biotechnol., 2012, vol. 23, no. 5, pp. 631–640. https://doi.org/10.1016/j.copbio.2011.12.025
Eggeling, L. and Bott, M., A giant market and a powerful metabolism: L-lysine provided by Corynebacterium glutamicum,Appl. Microbiol. Biotechnol., 2015, vol. 99, no. 8, pp. 3387–3394. https://doi.org/10.1007/s00253-015-6508-2
Becker, J., Giebelmann, G., Hoffmann, S.L., and Wittmann, C., Corynebacterium glutamicum for sustainable bioproduction: from metabolic physiology to systems metabolic engineering, Adv. Biochem. Eng. Biotechnol., 2018, vol. 162, pp. 217–263. https://doi.org/10.1007/10_2016_21
Moritz, B., Striegel, K., de Graaf, A.A., and Sahm, H., Kinetic properties of the glucose-6-phosphate and 6‑phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo, Eur. J. Biochem., 2000, vol. 267, no. 12, pp. 3442–3452. https://doi.org/10.1046/j.1432-1327.2000.01354.x
Eikmanns, B.J., Rittmann, D., and Sahm, H., Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme, J. Bacteriol., 1995, vol. 177, no. 3, pp. 774–782. https://doi.org/10.1128/jb.177.3.774-782.1995
Gourdon, P., Baucher, M.F., Lindley, N.D., and Guyonvarch, A., Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism, Appl. Environ. Microbiol., 2000, vol. 66, no. 7, pp. 2981–2987. https://doi.org/10.1128/aem.66.7.2981-2987.2000
Marx, A., de Graaf, A.A., Wiechert, W., et al., Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing, Biotechnol. Bioeng., 1996, vol. 49, no. 2, pp. 111–129. https://doi.org/10.1002/(SICI)1097-0290(19960120)49:2<111::AID-BIT1>3.0.CO;2-T
Takeno, S., Murata, R., Kobayashi, R., Mitsuhashi, S., and Ikeda, M., Engineering of Corynebacterium glutamicum with an NADPH-generating glycolytic pathway for L-lysine production, Appl. Environ. Microbiol., 2010, vol. 76, no. 21, pp. 7154–7160. https://doi.org/10.1128/AEM.01464-10
Bommareddy, R.R., Chen, Z., Rappert, S., and Zeng, A.P., A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase, Metab. Eng., 2014, vol. 25, pp. 30–37. https://doi.org/10.1016/j.ymben.2014.06.005
Sauer, U., Canonaco, F., Heri, S., Perrenoud, A., and Fischer, E., The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli,J. Biol. Chem., 2004, vol. 279, no. 8, pp. 6613–6619. https://doi.org/10.1074/jbc.M311657200
Kabus, A., Georgi, T., Wendisch, V.F., and Bott, M., Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves L-lysine formation, Appl. Microbiol. Biotechnol., 2007, vol. 75, no. 1, pp. 47–53. https://doi.org/10.1007/s00253-006-0804-9
Witthoff, S., Eggeling, L., Bott, M., and Polen, T., Corynebacterium glutamicum harbours a molybdenum cofactor-dependent formate dehydrogenase which alleviates growth inhibition in the presence of formate, Microbiology, 2012, vol. 158, no. 9, pp. 2428–2439. https://doi.org/10.1099/mic.0.059196-0
Tishkov, V.I. and Popov, V.O., Catalytic mechanism and application of formate dehydrogenase, Biochemistry (Moscow), 2004, vol. 69, no. 11, pp. 1252–1267.
Alekseeva, A.A., Fedorchuk, V.V., Zarubina, S.A., et al., The role of Ala198 in the stability and coenzyme specificity of bacterial formate dehydrogenases, Acta Nat., 2015, vol. 7, no. 1, pp. 60–69.
Ryabchenko, L.E., Shustikova, T.E., Sheremet’eva, M.E., Gerasimova, T.V., L-lysine-producing coryneform bacteria with an inactivated ltbR gene and a method for producing L-lysine using this bacterium, RF Patent No. 2639247 C1.
Tarutina, M.G., Raevskaya, N.M., Shustikova, T.E., et al., Assessment of effectiveness of Corynebacterium glutamicum promoters and their application for the enhancement of gene activity in lysine-producing bacteria, Appl. Biochem. Microbiol., 2016, vol. 52, no. 7, pp. 692–698.
Green, M.R. and Sambrook, J., Molecular Cloning: A Laboratory Manual, 4th ed., New York: Cold Spring Harbor Laboratory Press, 2011.
Van der Rest, M.E., Lange, C., and Molenaar, D., A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA, Appl. Microbiol. Biotechnol., 1999, vol. 52, no. 4, pp. 541–545. https://doi.org/10.1007/s002530051557
Hoelsch, K., Suhrer, I., Heusel, M., et al., Engineering of formate dehydrogenase: synergistic effect of mutations affecting cofactor specificity and chemical stability, Appl. Microbiol. Biotechnol., 2013, vol. 97, pp. 2473–2481. https://doi.org/10.1007/s00253-012-4142-9
ACKNOWLEDGMENTS
This work was carried out on the equipment of the Multipurpose Scientific Installation of All-Russia Collection of Industrial Microorganisms of the Kurchatov Institute National Resource Center (GOSNIIGENETIKA).
Funding
This work was financially supported by the Ministry of Education and Science of Russia (Unique Project Identifier RFMEFI61017X0011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This article does not contain any studies involving animals performed by any of the authors.
This article does not contain any studies involving human participants performed by any of the authors.
Additional information
Translated by I. Gordon
Abbreviations: CL—culture liquid; FDH–formate dehydrogenase; NADPH—nicotinamide-β-adenine dinucleotide phosphate, reduced; OD—optical density; PdeFDH_D221Q—NADPH+-dependent FDH with the D221Q mutation.
Rights and permissions
About this article
Cite this article
Ryabchenko, L.E., Leonova, T.E., Shustikova, T.E. et al. Expression of the NADPH+-Dependent Formate-Dehydrogenase Gene from Pseudomonas Increases Lysine Production in Corynebacterium glutamicum . Appl Biochem Microbiol 56, 828–836 (2020). https://doi.org/10.1134/S0003683820080086
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683820080086