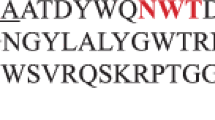Abstract
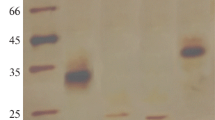
The heterologous expression and characteristics of a new xylanase from Pyromyces finnis are described. The endo-1,4-β-xylanase XylP (EC 3.2.1.8) consists of 223 amino acids and 19 residues of a putative signal peptide in the N-terminal region. The amino-acid sequence of the mature protein has the greatest homology (84%) with the sequence of the native catalytic N-terminal domain of Neocallimastix patriciarum endo-1,4-β-xylanase. A synthetic nucleotide sequence encoding the mature XylP protein was expressed in Pichia pastoris. The purified recombinant enzyme was active with birch xylan and arabinoxylan as substrates. The optimal pH and temperature for enzyme activity were established as 5.0 and 50°C, respectively, with the use of birch xylan. The specific activity of xylanase was 4700 U/mg protein, and KM and Vmax were equal to 0.51 mg/mL and 7395.3 μmol/(min mg), respectively. The recombinant XylP protein showed moderate thermal and high pH stability, as well as resistance to digestive enzymes and protein xylanase inhibitors from cereals. It was also shown that Mg2+, Co2+ and Li+ ions have a positive effect on enzyme activity.





Similar content being viewed by others
REFERENCES
Prade, R.A., Xylanases: from biology to biotechnology, Biotechnol. Genet. Eng. Rev., 1996, vol. 13, pp. 101–131.
Kulkarni, N., Shendye, A., and Rao, M., Molecular and biotechnological aspects of xylanases, FEMS Microbiol. Rev., 1999, vol. 23, no. 4, pp. 411–456.
Liab, K., Azadi, P., Collins, R., et al., Relationships between activities of xylanases and xylan structures, Enzyme Microbiol. Technol., 2000, vol. 27, nos. 1–2, pp. 89–94. https://doi.org/10.1016/S0141-0229(00)00190-3
Prade, R.A., Xylanases: from biology to biotechnology, Biotechnol. Genet. Eng. Rev., 1996, vol. 13, no. 1, pp. 101–132.
Manji, A.H., Extended usage of xylanase enzyme to enhance the bleaching of softwood kraft pulp, Tappi J., 2006, vol. 5, no. 1, pp. 23–26.
Coughlan, M.P. and Hazlewood, G.P., Hemicellulose and Hemicellulases, Portland: Portland Press, 1993.
Collins, T., Gerday, C., and Feller, G., Xylanases, xylanase families and extremophilic xylanases, FEMS Microbiol. Rev., 2005, vol. vol. 29, no. 1, pp. 3–23. https://doi.org/10.1016/j.femsre.2004.06.005
Gusakov, A.V., Protein inhibitors of microbial xylanases. Overview, Biochemistry (Moscow), 2010, vol. 75, no. 10, pp. 1185–1199. https://doi.org/10.1134/S0006297910100019
Lee, S.S., Ha, J.K., and Cheng, K.J., Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions, Appl. Environ. Microbiol., 2000, vol. 66, no. 9, pp. 3807–3813. https://doi.org/10.1128/AEM.66.9.3807-3813.2000
Orpin, C.G., Studies on the rumen flagellate Neocallimastix frontalis,Microbiology, 1975, vol. 91, no. 2, pp. 249–262.
Qiu, X., Selinger, B., Yanke, L.J., and Cheng, K.J., Isolation and analysis of two cellulase cDNAs from Orpinomyces joyonii,Gene, 2000, vol. 245, no. 1, pp. 119–126. https://doi.org/10.1016/S0378-1119(00)00028-7
Comlekcioglu, U., Akyol, I., Ozkose, E., et al., Carboxymethylcellulase production by the anaerobic rumen fungus Neocallimastix sp. GMLF7, Ann. Microbiol., 2008, vol. 58, no. 1, pp. 115–119.
Tsai, C.F., Qiu, X., and Liu, J.H., A comparative analysis of two cDNA clones of the cellulase gene family from anaerobic fungus Piromyces rhizinflata,Anaerobe, 2003, vol. 9, no. 3, pp. 131–140. https://doi.org/10.1016/S1075-9964(03)00087-8
Youssef, N.H., Couger, M.B., Struchtemeyer, C.G., et al., The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader, Appl. Environ. Microbiol., 2013, vol. 79, no. 15, pp. 4620–4634. https://doi.org/10.1128/AEM.00821-13
Harhangi, H.R., Steenbakkers, P.J., Akhmanova, A., Jetten, M.S., van der Drift, C., and Camp, H.J.O., A highly expressed family 1 β-glucosidase with transglycosylation capacity from the anaerobic fungus Piromyces sp. E2, Biochim. Biophys. Acta—Gene Struct.Expr., 2002, vol. 1574, no. 3, pp. 293–303. https://doi.org/10.1016/S0167-4781(01)00380-3
Ljungdahl, L.G., The cellulase/hemicellulase system of the anaerobic fungus Orpinomyces PC2 and aspects of its applied use, Ann. N.Y. Acad. Sci., 2008, vol. 1125, no. 1, pp. 308–321. https://doi.org/10.1196/annals.1419.030
Borneman, W.S., Hartley, R.D., Morrison, W.H., et al., Feruloyl and p-coumaroyl esterase from anaerobic fungi in relation to plant cell wall degradation, Appl. Microbiol. Biotechnol., 1990, vol. 33, no. 3, pp. 345–351. https://doi.org/10.1007/BF00164534
Borneman, W.S., Akin, D.E., and Ljungdahl, L.G., Fermentation products and plant cell wall-degrading enzymes produced by monocentric and polycentric anaerobic ruminal fungi, Appl. Environ. Microbiol., 1989, vol. 55, no. 5, pp. 1066–1073.
Li, X.L., Skory, C.D., Ximenes, E.A., et al., Expression of an AT-rich xylanase gene from the anaerobic fungus Orpinomyces sp. strain PC-2 in and secretion of the heterologous enzyme by Hypocrea jecorina,Appl. Microbiol. Biotechnol., 2007, vol. 74, no. 6, pp. 1264–1275. https://doi.org/10.1007/s00253-006-0787-6
Tsai, C.T. and Huang, C.T., Overexpression of the Neocallimastix frontalis xylanase gene in the methylotrophic yeasts Pichia pastoris and Pichia methanolica,Enzyme Microb. Technol., 2008, vol. 42, no. 6, pp. 459–465. https://doi.org/10.1016/j.enzmictec.2008.01.018
Van Wyk, N., Den, HaanR., and Van Zyl, W.H., Heterologous production of NpCel6A from Neocallimastix patriciarum in Saccharomyces cerevisiae,Enzyme Microb. Technol., 2010, vol. 46, no. 5, pp. 378–383. https://doi.org/10.1016/j.enzmictec.2009.11.005
O'Malley, M.A., Theodorou, M.K., and Kaiser, C.A., Evaluating expression and catalytic activity of anaerobic fungal fibrolytic enzymes native Topiromyces sp. E2 in Saccharomyces cerevisiae,Environ. Progr. Sustain. Energy, 2012, vol. 31, no. 1, pp. 37–46. https://doi.org/10.1002/ep.10614
Gordeeva, T.L., Borschevskaya, L.N., and Sineoky, S.P., Improved PCR-based gene synthesis method and its application to the Citrobacter freundii phytase gene codon modification, J. Microbiol. Methods, 2010, vol. 81, no. 2, pp. 147–152.
Kalinina, A.N., Gordeeva, T.L., and Sineoky, S.P., Expression of the xylanase gene from Paenibacillus brasilensis X1 in Pichia pastoris and characterization of the recombinant protein, Biotekhnologiya, 2018, vol. 34, no. 6, pp. 22–32. https://doi.org/10.21519/0234-2758-2018-34-6-22-32
Yurkevich, V.V., Practical Work on Biochemistry, Moscow: MSU, 1979, pp. 159–175.
Miller, G.L., Use of dinitrosalicylic acid reagent for determination of reducing sugar, Anal. Chem., 1959, vol. 31, no. 3, pp. 426–428.
Cheng, Y.S., Chen, C.C., Huang, C.H., et al., Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum insights into the molecular basis of a thermophilic enzyme, J. Biol. Chem., 2014, vol. 289, no. 16, pp. 11020–11028. https://doi.org/10.1074/jbc.M114.550905
Cereghino, J.L. and Cregg, J.M., Heterologous protein expression in the methylotrophic yeast Pichia pastoris,FEMS Microbiol. Rev., 2000, vol. 24, no. 1, pp. 45–66. https://doi.org/10.1111/j.1574-6976.2000.tb00532.x
Funding
The work was performed with the financial support of the Ministry of Education and Science of Russia (Unique Project Identifier RFMEFI60717X0180) with the Unique Scientific Installation, the Kurchatov Institute National Resource Center of the All-Russia Collection of Industrial Microorganisms (Kurchatov Institute NRC, GOSNIIGENETIKA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
This article does not contain any studies involving animals or humans performed by any of the authors.
Additional information
Translated by I. Gordon
Abbreviations: aa—amino-acid residue(s); CL—culture liquid; CMC—carboxymethylcellulose; DNS—3,5-dinitrosalicilic acid; DNS method—method to assess xylanase activity with dinitrosalicylic acid; EDTA—ethylendiamine tetraacetate; KM—Michaelis constant; PAGE—polyacrylamide gel electrophoresis; PCR—polymerase chain reaction; SDS—sodium dodecy(lauryl)sulfate; Vmax—xylanase maximum reaction rate.
Rights and permissions
About this article
Cite this article
Kalinina, A.N., Borshchevskaya, L.N., Gordeeva, T.L. et al. Expression of the Xylanase Gene from Pyromyces finnis in Pichia pastoris and Characterization of the Recombinant Protein. Appl Biochem Microbiol 56, 787–793 (2020). https://doi.org/10.1134/S0003683820070054
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0003683820070054