Anti-Allergic Potential of Cinnamaldehyde via the Inhibitory Effect of Histidine Decarboxylase (HDC) Producing Klebsiella pneumonia
Abstract
:1. Introduction
2. Results
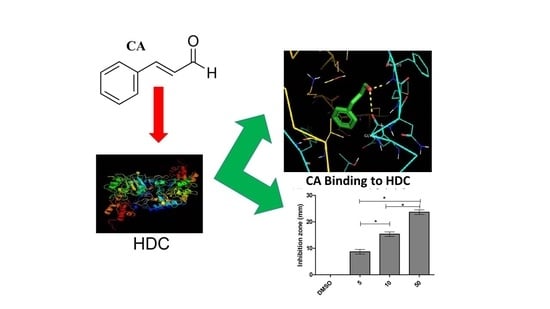
2.1. Structural and Docking Study of the HDC Protein
2.2. Inhibition of Bacterial HDC Activity by CA
2.3. CA Cytoprotective Role against IgE-Mediated Response and HDC Activity
2.4. CA Attenuates Antigen Induced Mast Cell Response
2.5. Modulatory Effects of Pro-Inflammatory Mediators
2.6. Effects of CA on In Vitro Allergic Markers
3. Discussion
4. Materials and Methods
4.1. Computation of Primary and Secondary Parameters of HDC Protein
4.2. The Prediction and Evaluation of the Three-Dimensional Model
4.3. Molecular Docking of Cinnamaldehyde
4.4. An Assay of HDC Activity
4.5. Cell Viability and Cytotoxic Study Using Stimulated RBL-2H3 Cells
4.6. An Assay of Inhibitory Effects of HDC
4.7. Determination of Nuclear Integrity, Cell Granulation and Apoptosis
4.8. Quantification of Nitric Oxide (NO) and Immunoblot
4.9. Analysis by Real-Time PCR
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pawankar, R. Allergic diseases and asthma: A global public health concern and a call to action. World Allergy Organ. J. 2014, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascal, M.; Perez-Gordo, M.; Caballero, T.; Escribese, M.M.; Lopez Longo, M.N.; Luengo, O.; Manso, L.; Matheu, V.; Seoane, E.; Zamorano, M.; et al. Microbiome and Allergic Diseases. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Cortes, A.; Castillo, Á.; Sciaraffia, A. Development of the scale of psychosocial factors in food allergy (SPS-FA). Pediatr. Allergy Immunol. 2013, 24, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, N. Expression of Histidine Decarboxylase and Its Roles in Inflammation. Int. J. Mol. Sci. 2019, 20, 376. [Google Scholar] [CrossRef] [Green Version]
- Ohtsu, H.; Tanaka, S.; Terui, T.; Hori, Y.; Makabe-Kobayashi, Y.; Pejler, G.; Tchougounova, E.; Hellman, L.; Gertsenstein, M.; Hirasawa, N.; et al. Mice lacking histidine decarboxylase exhibit abnormal mast cells. FEBS Lett. 2001, 502, 53–56. [Google Scholar] [CrossRef] [Green Version]
- Shade, K.T.C.; Conroy, M.E.; Washburn, N.; Kitaoka, M.; Huynh, D.J.; Laprise, E.; Patil, S.U.; Shreffler, W.G.; Anthony, R.M. Sialylation of immunoglobulin E is a determinant of allergic pathogenicity. Nature 2020, 582, 265–270. [Google Scholar] [CrossRef]
- Coleman, J.W. Nitric oxide: A regulator of mast cell activation and mast cell-mediated inflammation. Clin. Exp. Immunol. 2002, 129, 4–10. [Google Scholar] [CrossRef]
- Wershil, B.K.; Wang, Z.S.; Gordon, J.R.; Galli, S.J. Recruitment of neutrophils during IgE-dependent cutaneous late phase reactions in the mouse is mast cell-dependent. Partial inhibition of the reaction with antiserum against tumor necrosis factor-alpha. J. Clin. Investig. 1991, 87, 446–453. [Google Scholar] [CrossRef] [Green Version]
- Bidri, M.; Féger, F.; Varadaradjalou, S.; Ben Hamouda, N.; Guillosson, J.-J.; Arock, M. Mast cells as a source and target for nitric oxide. Int. Immunopharmacol. 2001, 1, 1543–1558. [Google Scholar] [CrossRef]
- Krystel-Whittemore, M.; Dileepan, K.N.; Wood, J.G. Mast Cell: A Multi-Functional Master Cell. Front Immunol. 2016, 6, 620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salamon, P.; Shoham, N.G.; Gavrieli, R.; Wolach, B.; Mekori, Y.A. Human mast cells release Interleukin-8 and induce neutrophil chemotaxis on contact with activated T cells. Allergy 2005, 60, 1316–1319. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Huang, C.H.; Hu, S.; Peng, H.L.; Wu, S.J. Topical Spilanthol Inhibits MAPK Signaling and Ameliorates Allergic Inflammation in DNCB-Induced Atopic Dermatitis in Mice. Int. J. Mol. Sci. 2019, 20, 2490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senthil, K.J.; Hsieh, H.W.; Wang, S.Y. Anti-inflammatory effect of lucidone in mice via inhibition of NF-κB/MAP kinase pathway. Int. Immunopharmacol. 2010, 10, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Babina, M.; Wang, Z.; Franke, K.; Guhl, S.; Artuc, M.; Zuberbier, T. Yin-Yang. Of IL-33 in Human Skin Mast Cells: Reduced Degranulation, but Augmented Histamine Synthesis through p38 Activation. J. Invest. Dermatol. 2019, 139, 1516–1525.e3. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Li, Y.; Liang, J.; Finkelman, F.D. Molecular Regulation of Histamine Synthesis. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Hirasawa, N.; Ohtsu, H.; Watanabe, T.; Ohuchi, K. Enhancement of neutrophil infiltration in histidine decarboxylase-deficient mice. Immunology 2002, 107, 217–221. [Google Scholar] [CrossRef]
- Ose, R.; Tu, J.; Schink, A.; Maxeiner, J.; Schuster, P.; Lucas, K.; Saloga, J.; Bellinghausen, I. Cinnamon extract inhibits allergen-specific immune responses in human and murine allergy models. Clin. Exp. Allergy 2019, 50, 41–50. [Google Scholar] [CrossRef]
- Nitta, Y.; Kikuzaki, H.; Ueno, H. Food Components Inhibiting Recombinant Human Histidine Decarboxylase Activity. J. Agric. Food Chem. 2007, 55, 299–304. [Google Scholar] [CrossRef]
- Hanieh, H.; Islam, V.I.H.; Saravanan, S.; Chellappandian, M.; Ragul, K.; Durga, A.; Venugopal, K.; Senthilkumar, V.; Senthilkumar, P.; Thirugnanasambantham, K. Pinocembrin, a novel histidine decarboxylase inhibitor with anti-allergic potential in in vitro. Eur. J. Pharmacol. 2017, 814, 178–186. [Google Scholar] [CrossRef]
- Gill, A.O.; Holley, R.A. Mechanisms of bactericidal action of cinnamaldehyde against Listeria monocytogenes and of eugenol against L. monocytogenes and Lactobacillus sakei. Appl Environ Microbiol. 2004, 70, 5750–5755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wendakoon, C.N.; Sakaguchi, M. Inhibition of Amino Acid Decarboxylase Activity of Enterobacter aerogenes by Active Components in Spices. J. Food Prot. 1995, 58, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.E.; Na, J.Y.; Lee, J.S. Anti-inflammatory effects of trans-cinnamaldehyde on lipopolysaccharide-stimulated macrophage activation via MAPKs pathway regulation. Immunopharmacol. Immunotoxicol. 2018, 40, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Gao, R.; Zhou, G.; Liu, J.; Li, J.; Shen, J. Trans-Cinnamaldehyde Inhibits IL-1β-Stimulated Inflammation in Chondrocytes by Suppressing NF-κB and p38-JNK Pathways and Exerts Chondrocyte Protective Effects in a Rat Model of Osteoarthritis. BioMed Res. Int. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özoğul, F. Production of biogenic amines by Morganella morganii, Klebsiella pneumoniae and Hafnia alvei using a rapid HPLC method. Eur. Food Res. Technol. 2004, 219, 465–469. [Google Scholar] [CrossRef]
- Guzman, J.D. Natural cinnamic acids, synthetic derivatives and hybrids with antimicrobial activity. Molecules 2014, 25, 19292–19349. [Google Scholar] [CrossRef]
- Dhara, L.; Tripathi, A. Antimicrobial activity of eugenol and cinnamaldehyde against extended spectrum beta lactamase producing enterobacteriaceae by in vitro and molecular docking analysis. Eur. J. Integr. Med. 2013, 5, 527–536. [Google Scholar] [CrossRef]
- Mateen, S.; Rehman, T.; Shahzad, S.; Naeem, S.S.; Faizy, A.F.; Khan, A.Q.; Khan, M.S.; Husain, F.M.; Moin, S. Anti-oxidant and anti-inflammatory effects of cinnamaldehyde and eugenol on mononuclear cells of rheumatoid arthritis patients. Eur. J. Pharmacol. 2019, 852, 14–24. [Google Scholar] [CrossRef]
- Walanj, S.; Walanj, A.; Mohan, V.; Thakurdesai, P.A. Efficacy and safety of the topical use of intranasal cinnamon bark extract in seasonal allergic rhinitis patients: A double-blind placebo-controlled pilot study. J. Herb. Med. 2014, 4, 37–47. [Google Scholar] [CrossRef]
- Thirugnanasambantham, K.; Muralidaran, S.; Mandal, A.K.A. Molecular Cloning, Computational and Expression Analysis of Anthocyanidin Reductase in Tea (Camellia sinensis). Appl. Biochem. Biotechnol. 2014, 174, 130–145. [Google Scholar] [CrossRef]
- Saravanan, S.; Islam, V.I.H.; Babu, N.P.; Pandikumar, P.; Thirugnanasambantham, K.; Chellappandian, M.; Raj, C.S.D.; Paulraj, M.G.; Ignacimuthu, S. Swertiamarin attenuates inflammation mediators via modulating NF-κB/I κB and JAK2/STAT3 transcription factors in adjuvant induced arthritis. Eur. J. Pharm. Sci. 2014, 56, 70–86. [Google Scholar] [CrossRef]
- Baker, D. Protein Structure Prediction and Structural Genomics. Science 2001, 294, 93–96. [Google Scholar] [CrossRef] [Green Version]
- Moya-García, A.A.; Pino-Ángeles, A.; Gil-Redondo, R.; Morreale, A.; Sánchez-Jiménez, F. Structural features of mammalian histidine decarboxylase reveal the basis for specific inhibition. Br. J. Pharmacol. 2009, 157, 4–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandmeier, E.; Hale, T.I.; Christen, P. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 1994, 221, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Melgarejo, E.; Medina, M.Á.; Sánchez-Jiménez, F.; Botana, L.M.; Dominguez, M.; Escribano, L.; Orfao, A.; Urdiales, J.L. (—)-Epigallocatechin-3-gallate interferes with mast cell adhesiveness, migration and its potential to recruit monocytes. Cell. Mol. Life Sci. 2007, 64, 2690–2701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Caso, C.; Rodriguez-Agudo, D.; Sánchez-Jiménez, F.; Medina, M.A. Green tea epigallocatechin-3-gallate is an inhibitor of mammalian histidine decarboxylase. Cell. Mol. Life Sci. 2003, 60, 1760–1763. [Google Scholar] [CrossRef] [PubMed]
- Moya-García, A.A.; Ruiz-Pernía, J.; Martí, S.; Sánchez-Jiménez, F.; Tuñón, I. Analysis of the Decarboxylation Step in Mammalian Histidine Decarboxylase. J. Boil. Chem. 2008, 283, 12393–12401. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Caso, C.; Rodríguez-Agudo, D.; Moya-García, A.A.; Fajardo, I.; Medina, M.Á.; Subramaniam, V.; Sánchez-Jiménez, F. Local changes in the catalytic site of mammalian histidine decarboxylase can affect its global conformation and stability. Eur. J. Biochem. 2003, 270, 4376–4387. [Google Scholar] [CrossRef] [Green Version]
- Landete, J.M.; Pardo, I.; Ferrer, S.; Landete, J. Histamine, histidine, and growth-phase mediated regulation of the histidine decarboxylase gene in lactic acid bacteria isolated from wine. FEMS Microbiol. Lett. 2006, 260, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Kageyama-Yahara, N.; Wang, X.; Katagiri, T.; Wang, P.; Yamamoto, T.; Tominaga, M.; Kadowaki, M. Suppression of phospholipase Cγ1 phosphorylation by cinnamaldehyde inhibits antigen-induced extracellular calcium influx and degranulation in mucosal mast cells. Biochem. Biophys. Res. Commun. 2011, 416, 283–288. [Google Scholar] [CrossRef]
- Hagenlocher, Y.; Kiessling, K.; Schäffer, M.; Bischoff, S.C.; Lorentz, A. Cinnamaldehyde is the main mediator of cinnamon extract in mast cell inhibition. Eur. J. Nutr. 2015, 54, 1297–1309. [Google Scholar] [CrossRef]
- Li, D.; Carozza, R.B.; Shatos, M.A.; Hodges, R.R.; Dartt, D.A. Co-Effect of Histamine on Ca2+-Dependent Signaling Pathways in Rat Conjunctival Goblet Cells. Investig. Ophthalmol. Vis. Sci. 2012, 53, 6928–6938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhakal, H.; Yang, E.-J.; Lee, S.; Kim, M.-J.; Baek, M.-C.; Lee, B.; Park, P.-H.; Kwon, T.K.; Khang, D.; Song, K.-S.; et al. Avenanthramide C from germinated oats exhibits anti-allergic inflammatory effects in mast cells. Sci. Rep. 2019, 9, 6884. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.-H.; Kim, S.-A. 2-Methoxycinnamaldehyde inhibits the TNF-α-induced proliferation and migration of human aortic smooth muscle cells. Int. J. Mol. Med. 2016, 39, 191–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsias, D.I.; Savvatianos, S.; Papadopoulos, N.G. An insight into the early mechanisms of allergen-specific immunotherapy. Immunotherapy 2011, 3, 333–336. [Google Scholar] [CrossRef]
- Yang, T.; Li, Y.; Lyu, Z.; Huang, K.; Corrigan, C.J.; Ying, S.; Wang, W.; Wang, C. Characteristics of Proinflammatory Cytokines and Chemokines in Airways of Asthmatics. Chin. Med. J. 2017, 130, 2033–2040. [Google Scholar] [CrossRef]
- Roth-Walter, F.; Moskovskich, A.; Gomez-Casado, C.; Diaz-Perales, A.; Oida, K.; Singer, J.; Kinaciyan, T.; Fuchs, H.C.; Jensen-Jarolim, E. Immune Suppressive Effect of Cinnamaldehyde Due to Inhibition of Proliferation and Induction of Apoptosis in Immune Cells: Implications in Cancer. PLoS ONE 2014, 9, e108402. [Google Scholar] [CrossRef]
- Boudreau, R.T.M.; Hoskin, D.W.; Lin, T.-J. Phosphatase inhibition potentiates IL-6 production by mast cells in response to FcεRI-mediated activation: Involvement of p38 MAPK. J. Leukoc. Boil. 2004, 76, 1075–1081. [Google Scholar] [CrossRef]
- Forsythe, P.; Gilchrist, M.; Kulka, M.; Befus, A.D. Mast cells and nitric oxide: Control of production, mechanisms of response Int. Immunopharmacology 2001, 1, 1525–1541. [Google Scholar] [CrossRef]
- Yatera, K.; Mukae, H. Possible pathogenic roles of nitric oxide in asthma. Respir. Investig. 2019, 57, 295–297. [Google Scholar] [CrossRef]
- Allen, J.B.; Wong, H.L.; Costa, G.L.; Bienkowski, M.J.; Wahl, S.M. Suppression of monocyte function and differential regulation of IL-1 and IL-1ra by IL-4 contribute to resolution of experimental arthritis. J. Immunol. 1993, 151, 4344–4351. [Google Scholar]
- Yorimitsu, M.; Nishida, K.; Shimizu, A.; Doi, H.; Miyazawa, S.; Komiyama, T.; Nasu, Y.; Yoshida, A.; Watanabe, S.; Ozaki, T. Intra-articular injection of interleukin-4 decreases nitric oxide production by chondrocytes and ameliorates subsequent destruction of cartilage in instability-induced osteoarthritis in rat knee joints. Osteoarthr. Cartil. 2008, 16, 764–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulker, O.C.; Ates, I.; Atak, A.; Karakaya, A. Evaluation of non-radioactive endpoints of ex vivo local lymph node assay-BrdU to investigate select contact sensitizers. J. Immunotoxicol. 2012, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kedong, H.; Wang, D.; Sagaram, M.; An, H.S.; Chee, A. Anti-inflammatory effects of interleukin-4 on intervertebral disc cells. Spine, J. 2019, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salamon, P.; Shefler, I.; Hershko, A.Y.; Mekori, Y.A. The Involvement of Protein Kinase D in T Cell-Induced Mast Cell Activation. Int. Arch. Allergy Immunol. 2016, 171, 203–208. [Google Scholar] [CrossRef] [Green Version]
- Yahfoufi, N.; Alsadi, N.; Jambi, M.; Matar, C. The Immunomodulatory and Anti-Inflammatory Role of Polyphenols. Nutrients 2018, 10, 1618. [Google Scholar] [CrossRef] [Green Version]
- Ahn, S.; Kim, E.; Lee, K.; Lee, D.-C. Cinnamaldehyde derivatives inhibit degranulation and inflammatory mediator production in rat basophilic leukemia cells. Int. Immunopharmacol. 2016, 38, 342–348. [Google Scholar] [CrossRef]
- Liu, W.; Liang, Q.; Balzar, S.; Wenzel, S.; Gorska, M.; Alam, R. Cell-specific activation profile of extracellular signal-regulated kinase 1/2, Jun N-terminal kinase, and p38 mitogen-activated protein kinases in asthmatic airways. J. Allergy Clin. Immunol. 2008, 121, 893–902.e2. [Google Scholar] [CrossRef]
- Kumar, A.; Lnu, S.; Malya, R.; Barron, D.; Moore, J.; Corry, D.B.; Boriek, A.M. Mechanical stretch activates nuclear factor-kappaB, activator protein-1, and mitogen-activated protein kinases in lung parenchyma: Implications in asthma. FASEB J. 2003, 17, 1800–1811. [Google Scholar] [CrossRef]
- Hancı, D.; Altun, H.; Çetinkaya, E.A.; Muluk, N.B.; Cengiz, B.P.; Cingi, C. Cinnamaldehyde is an effective anti-inflammatory agent for treatment of allergic rhinitis in a rat model. Int. J. Pediatr. Otorhinolaryngol. 2016, 84, 81–87. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Laurie, A.T.R.; Jackson, R.M. Q-SiteFinder: An energy-based method for the prediction of protein-ligand binding sites. Bioinformatics 2005, 21, 1908–1916. [Google Scholar] [CrossRef] [PubMed]
- Niven, C.F.; Jeffrey, M.B.; Corlett, D.A. Differential plating medium for quantitative detection of histamine-producing bacteria. Appl. Environ. Microbiol. 1981, 41, 321–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasul, A.; Khan, M.; Yu, B.; Ma, T.; Yang, H. Xanthoxyletin, a coumarin induces S phase arrest and apoptosis in human gastric adenocarcinoma SGC-7901 cells. Asian Pac. J. Cancer 2011, 12, 1219–1223. [Google Scholar]
- Kanki, M.; Yoda, T.; Tsukamoto, T.; Baba, E. Histidine Decarboxylases and Their Role in Accumulation of Histamine in Tuna and Dried Saury. Appl. Environ. Microbiol. 2007, 73, 1467–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, P.; Zarrouk, M.; Kawasaki, K.; Isoda, H. Inhibitory effect of various Tunisian olive oils on chemical mediator release and cytokine production by basophilic cells. J. Ethnopharmacol. 2008, 116, 279–287. [Google Scholar] [CrossRef]
- Kendrick, K.M.; Guevara-Guzman, R.; Zorrilla, J.; Hinton, M.R.; Broad, K.D.; Mimmack, M.; Ohkura, S. Formation of olfactory memories mediated by nitric oxide. Nature 1997, 388, 670–674. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |





| Primer Name | Forward | Reverse | Product Size |
|---|---|---|---|
| MAPK-6 | CCATCTCAAGCCACCCTTTCCA | CTCCGAGAACTGACAATCGTGG | 135 |
| ERK-1 | TACAAGCTTAGCTCGGCCTAT GACCACGTG | TACGAATTCGGCTTTAGATCT CGGTGGAGC | 264 |
| CXCL-8 | ATGACTTCCAAGCTGGCCGTGGCT | TCTCAGCCCTCTTCAAAAACTTCTC | 141 |
| COX-2 | GAATCATTCACCAGGCAAATTG | TCTGTACTGCGGGTGGAACA | 149 |
| β-ACTIN | GATGGCCACGGCTGCTTC | TGCCTCAGGGCAGCGGAA | 165 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badger-Emeka, L.I.; Emeka, P.M.; Thirugnanasambantham, K.; Ibrahim, H.I.M. Anti-Allergic Potential of Cinnamaldehyde via the Inhibitory Effect of Histidine Decarboxylase (HDC) Producing Klebsiella pneumonia. Molecules 2020, 25, 5580. https://doi.org/10.3390/molecules25235580
Badger-Emeka LI, Emeka PM, Thirugnanasambantham K, Ibrahim HIM. Anti-Allergic Potential of Cinnamaldehyde via the Inhibitory Effect of Histidine Decarboxylase (HDC) Producing Klebsiella pneumonia. Molecules. 2020; 25(23):5580. https://doi.org/10.3390/molecules25235580
Chicago/Turabian StyleBadger-Emeka, Lorina I., Promise Madu Emeka, Krishnaraj Thirugnanasambantham, and Hairul Islam M. Ibrahim. 2020. "Anti-Allergic Potential of Cinnamaldehyde via the Inhibitory Effect of Histidine Decarboxylase (HDC) Producing Klebsiella pneumonia" Molecules 25, no. 23: 5580. https://doi.org/10.3390/molecules25235580