Biosynthesis of Polyhydroxyalkanoates (PHAs) by the Valorization of Biomass and Synthetic Waste
Abstract
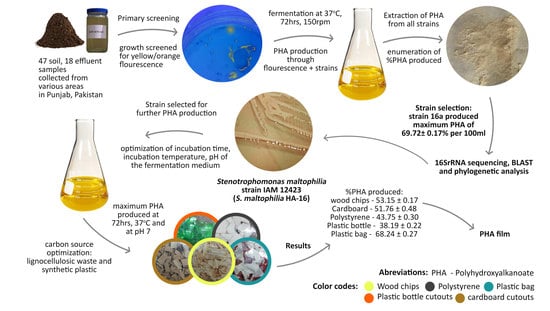
:1. Introduction
2. Results and Discussion
2.1. Primary Screening
2.2. Secondary Screening of PHA Producers
2.3. Molecular Identification
2.4. Characterization of the Extracted Polymer
2.5. Optimization of Cultural Conditions
2.5.1. Effect of Time of Incubation
2.5.2. Effect of Temperature of Incubation
2.5.3. Effect of pH of the Fermentation Medium
2.5.4. Optimization of Carbon Source for PHA Production
2.6. PHA Film Preparation
3. Materials and Methods
3.1. Reagents Preparation
3.1.1. Trace Elements Solutions
3.1.2. HCl (=35%) Solution Preparation
3.2. Media Preparation
3.2.1. PHA Detecting Agar
3.2.2. Nutrient Broth and Nutrient Agar
3.2.3. Fermentation Medium
3.2.4. Seed Culture Preparation
3.3. Sample Collection
3.4. Waste Collection
3.5. Isolation and Screening
3.6. Submerged Fermentation for PHA Production
3.7. Extraction of PHA Produced during Fermentation
3.8. Quantification of Produced PHA
3.9. Characterization of the Extracted PHA by FTIR
3.10. Molecular Identification of the Most Efficient PHA Producing Strain
3.11. Optimization of Cultural Conditions
3.12. PHA Film Preparation
3.13. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Global Plastics & Rubber Products Manufacturing Report. Available online: https://www.thebusinessresearchcompany.com/report/plastics-and-rubber-products-manufacturing-global-market-report (accessed on 1 September 2020).
- Grand View Research Inc. Plastics Market Size, Share & Trends Analysis Report By Product (PE, PP, PU, PVC, PET, Polystyrene, ABS, PBT, PPO, Epoxy Polymers, LCP, PC, Polyamide), By Application, and Segment Forecasts, 2019–2025; Grand View Research Inc., 2019; 200p, Available online: https://www.grandviewresearch.com/industry-analysis/global-plastics-market (accessed on 11 January 2020).
- Ecommerce Gateway Pakistan. Pakistan—The Regional Business Hub. Plastic, Packaging & Print Asia. In Proceedings of the 12th Plastic Machinery Technology Show, Karachi, Islamabad, and Lahore: Plastic, Packaging & Print Asia Secretariat, Karachi, Pakistan, 12–14 September 2020; E-commerce Gateway: Karachi, Pakistan, 2020. [Google Scholar]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and the fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plastic Pollution. Available online: https://coastalcare.org/2009/11/plastic-pollution/ (accessed on 2 February 2020).
- Ellen MacArthur Foundation. The New Plastics Economy: Catalysing Action; Ellen MacArthur Foundation and World Economic Forum: Isle of Wight, UK, 2017. [Google Scholar]
- Ebere, E.C.; Wirnkor, V.A.; Ngozi, V.E.; Chizoruo, I.F.; Collins, A. Airborne Microplastics: A Review Study on Method for Analysis, Occurrence, Movement, and Risks. PrePrints 2019, 191, 668. [Google Scholar] [CrossRef] [Green Version]
- Allen, S.; Allen, D.; Phoenix, V.R.; Roux, G.L.; Jiménez, P.D.; Simonneau, A.; Binet, S.; Galop, D. Atmospheric transport and deposition of microplastics in a remote mountain catchment. Nat. Geosci. 2019, 12, 339–344. [Google Scholar] [CrossRef]
- Nielson, T.D.; Hasselbalch, J.; Holmberg, K.; Stripple, J. Politics & Plastic crisis: A review throughout the plastic lifecycle. WIREs Energy Environ. 2020, 9, 360. [Google Scholar] [CrossRef] [Green Version]
- Ebnesajjad, S. Handbook of Biopolymers and Biodegradable Plastics: Properties, Processing, and Applications; Elsevier Inc.: Oxford, UK, 2013. [Google Scholar]
- Kaur, L.; Khajuria, R.; Parihar, L.; Singh, G.D. Polyhydroxyalkanoates: Biosynthesis to commercial production: A review. J. Microbiol. Biotechnol. Food Sci. 2017, 6, 1098–1106. [Google Scholar] [CrossRef] [Green Version]
- Koller, M. Advances in Polyhydroxyalkanoate (PHA) Production, Volume 2. Bioengineering 2020, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Parmar, N. Isolation and characterization of two novel polyhydroxybutyrate (PHB)-producing bacteria. Afr. J. Biotechnol. 2011, 10, 4907–4919. [Google Scholar]
- Goh, Y.S.; Tan, I.K.P. Polyhydroxyalkanoate production by antarctic soil bacteria isolated from Casey Station and Signy Island. Microbiol. Res. 2012, 167, 211–219. [Google Scholar] [CrossRef]
- Muhammadi; Shabina; Afzal, M.; Hameed, S. Bacterial polyhydroxyalkanoates-eco-friendly next generation plastic: Production, biocompatibility, biodegradation, physical properties and applications. Green Chem. Lett. Rev. 2015, 8, 56–77. [Google Scholar] [CrossRef] [Green Version]
- Paduani, M. Microplastics as novel sedimentary particles in coastal wetlands: A review. Mar. Pollut. Bull. 2020, 161, 111739. [Google Scholar] [CrossRef]
- Raza, Z.A.; Abid, S.; Banat, I.M. Polyhydroxyalkanoates: Characteristics, production, recent developments and applications. Int. Biodeterior. Biodegrad. 2018, 126, 45–56. [Google Scholar] [CrossRef]
- Cabrera, F.; Torres, Á.; Campos, J.L.; Jeison, D. Effect of Operational Conditions on the Behaviour and Associated Costs of Mixed Microbial Cultures for PHA Production. Polymers 2019, 11, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koller, M.; Maršálek, L.; Dias, M.D.S.; Braunegg, G. Producing microbial polyhydroxyalkanoate (PHA) biopolyesters in a sustainable manner. New Biotechnol. 2017, 37, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef] [Green Version]
- Plastic, Composition, Uses, Types and Facts, Britannica. Available online: https://www.britannica.com/science/plastic (accessed on 26 October 2020).
- Global Production Capacities of Bioplastics 2018–2023. Available online: www.european-bioplastics.org (accessed on 2 February 2020).
- Ciesielski, S.; Mozejko, J.; Przybylek, G. The influence of nitrogen limitation on mcl-PHA synthesis by two newly isolated strains of Pseudomonas sp. J. Ind. Microbiol. Biotechnol. 2010, 37, 511–520. [Google Scholar] [CrossRef]
- Doron, A. Waste of a Nation: Garbage and Growth in India; Harvard University Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Effluent Waste, InforMEA. Available online: https://www.informea.org/en/terms/effluent-waste#:~:text=Definition(s),of%20their%20polluting%20chemical%20composition.%20 (accessed on 26 October 2020).
- Koller, M.; Gasser, I.; Schmid, F.; Berg, G. Linking ecology with economy: Insights into polyhydroxyalkanoate-producing microorganisms. Eng. Life Sci. 2011, 11, 222–237. [Google Scholar] [CrossRef]
- Barnes, D.K.A.; Galgani, F.; Thompson, R.C.; Barlaz, M. Accumulation and fragmentation of plastic debris in global environments. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 1985–1998. [Google Scholar] [CrossRef] [Green Version]
- Ostle, G.A.; Holt, J.G. Nile Blue A as a Fluorescent Stain for Poly-3-Hydroxybutyrate. Appl. Environ. Microbiol. 1982, 44, 238–241. [Google Scholar] [CrossRef] [Green Version]
- Spiekermann, P.; Rehm, B.H.; Kalscheuer, R.; Baumeister, D.; Steinbüchel, A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 1999, 171, 73–80. [Google Scholar] [CrossRef]
- Mohammed, S.; Panda, A.N.; Ray, L. An investigation for recovery of polyhydroxyalkanoates (PHA) from Bacillus sp. BPPI-14 and Bacillus sp. BPPI-19 isolated from plastic waste landfill. Int. J. Biol. Macromol. 2019, 134, 1085–1096. [Google Scholar] [CrossRef]
- Kosseva, M.R.; Rusbandi, E. Trends in the biomanufacture of polyhydroxyalkanoates with focus on downstream processing. Int. J. Biol. Macromol. 2018, 107, 762–778. [Google Scholar] [CrossRef] [PubMed]
- Sangkharak, K.; Prasertsan, P. Screening and identificationof polyhydroxyalkanoates producing bacteria and bio-chemical characterization of their possible application. J. Gen. Appl. Microbiol. 2012, 58, 173–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shovitri, M.; Nafi’Ah, R.; Antika, T.R.; Alami, N.H.; Kuswytasari, N.D.; Zulaikha, E. Soil burial method for plastic degradation performed by Pseudomonas PL-01, Bacillus PL-01, and indigenous bacteria. AIP Conf. Proc. 2011, 1854, 020035. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Zhao, D.; Li, C. Isolation and 2,4-D-degrading characteristics of Cupriavidus campinensis BJ71. Braz. J. Microbiol. 2015, 46, 433–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puglisi, E.; Romaniello, F.; Galletti, S.; Boccaleri, E.; Frache, A.; Cocconcelli, P.S. Selective bacterial colonization processes on polyethylene waste samples in an abandoned landfill site. Sci. Rep. 2019, 9, 14138. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Schäffer, A.A.; Agarwala, R.; Altschul, S.F.; Lipman, D.J.; Madden, T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct 2012, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1089–1191. [Google Scholar] [CrossRef] [Green Version]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; Lelie, D.; Dow, M. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef]
- Gilligan, P.H.; Lum, G.; VanDamme, P.A.R.; Whittier, S. Burkholderia, Stenotrophomonas, Ralstonia, Brevundimonas, Comamonas, Delftia, Pandoraea, and Acidivorax. In Manual of Clinical Microbiology, 8th ed.; Murray, P.R., Baron, E.J., Jorgensen, J.H., Pfaller, M.A., Yolken, R.H., Eds.; ASM Press: Washington, DC, USA, 2003; pp. 729–748. [Google Scholar]
- Patil, P.P.; Midha, S.; Kumar, S.; Patil, P.B. Genome Sequence of Type Strains of Genus Stenotrophomonas. Front. Microbiol. 2016, 7, 309. [Google Scholar] [CrossRef] [Green Version]
- Horiike, T. An introduction to molecular phylogenetic analysis. Rev. Agric. Sci. 2016, 4, 36–45. [Google Scholar] [CrossRef] [Green Version]
- Ziemert, N.; Jensen, P.R. Phylogenetic approaches to natural product structure prediction. Methods Enzymol. 2015, 517, 161–182. [Google Scholar] [CrossRef] [Green Version]
- OriginLab Corporation. Origin (Pro), Version Number (e.g., “Version 8.5”); OriginLab Corporation: Northampton, MA, USA, 2010. [Google Scholar]
- Abid, S.; Raza, A.Z.; Hussain, T. Production kinetics of polyhydroxyalkanoates by using Pseudomonas aeruginosa gamma ray mutant strain EBN-8 cultured on soybean oil. 3 Biotech 2016, 6, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, K.C.; Pereira, P.; Smith, A.C.; Carvalho, C.W.P.; Wellner, N.; Yakimets, I. Characterization of polyhydroxybutyrate-hydroxyvalerate (PHB-HV)/maize starch blend films. J. Food Eng. 2008, 89, 361–369. [Google Scholar] [CrossRef]
- Pérez-Arauz, A.O.; Aguilar-Rabiela, A.E.; Vargas-Torres, A.; Rodríguez-Hernández, A.I.; Chavarría-Hernández, N.; Vergara-Porras, B.; López-Cuellar, M.R. Production and characterization of biodegradable films of a novel polyhydroxyalkanoate (PHA) synthesized from peanut oil. Food Packag. Shelf. Life 2019, 20, 100297. [Google Scholar] [CrossRef]
- Ribitsch, D.; Heumann, S.; Karl, W.; Gerlach, J.; Leber, R.; Birner-Gruenberger, R.; Gruber, K.; Eiteljoerg, I.; Remler, P.; Siegert, P.; et al. Extracellular serine proteases from Stenotrophomonas maltophilia: Screening, isolation and heterologous expression in E. coli. J. Biotechnol. 2012, 157, 140–147. [Google Scholar] [CrossRef]
- Khare, E.; Chopra, J.; Arora, N.K. Screening for mcl-PHAproducing fluorescent Pseudomonads and comparison of mclPHA production under iso-osmotic conditions induced by PEG and NaCl. Curr. Microbiol. 2014, 68, 457–462. [Google Scholar] [CrossRef]
- Munir, S.; Iqbal, S.; Jamil, N. Polyhydroxyalkanoates (PHA) production using paper mill wastewater as carbon source in comparison with glucose. J. Pure Appl. Microbiol. 2015, 9, 453–460. [Google Scholar]
- Alqahtani, N. The Effect of Varying Environmental Conditions on the Growth of Stenotrophomonas Maltophilia. Master’s Thesis, University of the Incarnate Word, San Antonio, TX, USA, 2016. [Google Scholar]
- Shaaban, M.T.; Mowafy, E.I. Studies on incubation periods, scale up and biodisintegration of poly-β-hydroxybutyrate (PHB) synthesis by Stenotrophomonas (pseudomonas) maltophilia and Pseudomonas putida. Egypt. J. Exp. Biol. (Bot.) 2012, 8, 133–140. [Google Scholar]
- Dai, D.; Reusch, R.N. Poly-3-hydroxybutyrate Synthase from the Periplasm of Escherichia coli. Biochem. Biophys. Res. Commun. 2008, 374, 485–489. [Google Scholar] [CrossRef] [Green Version]
- Guerrero, C.E.; De la Rosa, G.; Gonzalez Castañeda, J.; Sánchez, Y.; Castillo-Michel, H.; Valdez-Vazquez, I.; Balcazar, E.; Salmerón, I. Optimization of Culture Conditions for Production of Cellulase by Stenotrophomonas maltophilia. BioResources 2018, 13, 8358–8372. [Google Scholar] [CrossRef]
- Shaaban, M.T.; Attia, M.; Mowafy, E.I. Production of some biopolymers by some selective Egyptian soil bacterial isolates. J. Appl. Sci. Res. 2012, 8, 94–105. [Google Scholar]
- Raj, A.; Kumar, S.; Singh, S.K. A highly thermostable xylanase from Stenotrophomonas maltophilia: Purification and Characterization. Enzyme Res. 2013, 8, 429305. [Google Scholar] [CrossRef]
- Lathwal, P.; Nehra, K.; Singh, M.; Jamdagni, P.; Rana, J.S. Optimization of Culture Parameters for Maximum Polyhydroxybutyrate Production by Selected Bacterial Strains Isolated from Rhizospheric Soils. Pol. J. Microbiol. 2015, 64, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Morales-Jimenez, J.; Zuñiga, G.; Ramırez-Saad, H.C.; Hernandez-Rodrıguez, C. Gut-Associated Bacteria Throughout the Life Cycle of the Bark Beetle Dendroctonus rhizophagus (Curculionidae: Scolytinae) and Their Cellulolytic Activities. Microb. Ecol. 2012, 64, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Kirtania, K.; Tanner, J.; Kabir, K.; Rajendran, S.; Bhattacharya, S. In situ Snychrotron IR study relating temperature and heating rate to surface functional group changes in biomass. Bioresour. Technol. 2014, 151, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Ali Wala’a, S.; Zaki, H.N.; Obaid, S.Y.N. Production of bioplastic by bacteria isolated from local soil and organic wastes. Curr. Res. Microbiol. Biotechnol. 2017, 5, 1012–1017. [Google Scholar]
- Speight, J.G.; Lange, N.A. Lange’s Handbook of Chemistry, 16th ed.; McGraw-Hill: New York, NY, USA, 2005; pp. 2758–2807. [Google Scholar]
- Plastics Europe. Plastics: The Facts; Plastics Europe: Brussels, Belgium, 2018. [Google Scholar]
- Why Doesn’t Plastic Biodegrade? Live Science. Available online: https://www.livescience.com/33085-petroleum-derived-plastic-non-biodegradable.html (accessed on 27 October 2020).
- Kenny, S.T.; Runic, J.N.; Kaminsky, W.; Woods, T.; Babu, R.P.; Keely, C.M.; Blau, W.; O’Connor, K.E. Up-cycling of PET (Polyethylene Terephthalate) to the biodegradable plastic PHA (Polyhydroxyalkanoate). Environ. Sci. Technol. 2008, 42, 7696–7701. [Google Scholar] [CrossRef]
- Johnston, B.; Radecka, I.; Hill, D.; Chiellini, E.; Ilieva, V.I.; Sikorska, W.; Musioł, M.; Ziȩba, M.; Marek, A.A.; Keddie, D.; et al. The microbial production of Polyhydroxyalkanoates from Waste polystyrene fragments attained using oxidative degradation. Polymers (Basel) 2018, 10, 957. [Google Scholar] [CrossRef] [Green Version]
- Berg, G.; Eberl, L.; Hartmann, A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 2005, 7, 1673–1685. [Google Scholar] [CrossRef]
- Denet, E.; Coupat-Goutaland, B.; Nazaret, S.; Pelandakis, M.; Favre-Bonte, S. Diversity of free-living amoebae in soils and their associated human opportunistic bacteria. Parasitol. Res. 2017, 116, 3151–3162. [Google Scholar] [CrossRef] [Green Version]
- Somaraja, P.K.; Gayathri, D.; Ramaiah, N. Molecular Characterization of 2-Chlorobiphenyl Degrading Stenotrophomonas maltophilia GS-103. Bull. Environ. Contam. Toxicol. 2013, 91, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.J.; Shaikh, S.S.; Tabassum, B.; Thakur, R.; Gulati, A.; Sayyed, R.Z. Stenotrophomonas sp. RZS 7, a novel PHB degrader isolated from plastic contaminated soil in Shahada, Maharashtra, Western India. 3 Biotech 2016, 6, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, P.; Roy, P. Genomic Potential of Stenotrophomonas maltophilia in Bioremediation with an Assessment of Its Multifaceted Role in Our Environment. Front. Microbiol. 2016, 7, 967. [Google Scholar] [CrossRef] [PubMed]
- Gamal, R.F.; Abdelhady, H.M.; Khodair, T.A.; El-Tayeb, T.S.; Hassan, E.A.; Aboutaleb, K.A. Semi-scale produFction of PHAs from waste frying oil by Pseudomonas fluorescens S48. Braz. J. Microbiol. 2013, 44, 539–549. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.M. Wood waste—Carbon source for Polyhydroxyalkanoates (PHAs) production. Int. J. For. Wood Sci. 2017, 4, 36–40. [Google Scholar]
- Jorgensen, J.H.; Pfaller, M.A.; Carroll, K.C.; Funke, G.; Landry, M.L.; Richter, S.S.; Warnock, D.W. Manual of Clinical Microbiology, 11th ed.; American Society for Microbiology Press: Washington, DC, USA, 2015. [Google Scholar]
- Ren, Y.; Ling, C.; Hajnal, I.; Wu, Q.; Chen, G.Q. Construction of Halomonas bluephagenesis capable of high cell density growth for efficient PHA production. Appl. Microbiol. Biotechnol. 2018, 102, 4499–4510. [Google Scholar] [CrossRef]
- Fauzi, A.H.M.; Chua, A.S.M.; Yoon, L.W.; Nittami, T.; Yeoh, H.K. Enrichment of PHA-accumulators for sustainable PHA production from crude glycerol. Process Saf. Environ. Prot. 2019, 122, 200–208. [Google Scholar] [CrossRef]
- Desouky, S.E.; Abdel-rahman, M.A.; Azab, M.S.; Esmael, M.E. Batch and fed-batch production of polyhydroxyalkanoates from sugarcane molasses by Bacillus flexus Azu-A 2. J. Innov. Pharm. Biol. Sci. 2017, 4, 55–66. [Google Scholar]
- Bhuwal, A.K.; Singh, G.; Aggarwal, N.K.; Goyal, V.; Yadav, A. Isolation and screening of Polyhydroxyalkanoates producing bacteria from pulp, paper, and cardboard industry wastes. Int. J. Biomater. 2013, 2013, 752821. [Google Scholar] [CrossRef] [Green Version]
- PET Plastic. Available online: https://omnexus.specialchem.com/selection-guide/polyethylene-terephthalate-pet-plastic (accessed on 7 November 2020).
- Polyethylene Terephthalate. Available online: https://www.britannica.com/science/polyethylene-terephthalate (accessed on 7 November 2020).
- Polyethylene (PE) Plastic. Available online: https://omnexus.specialchem.com/selection-guide/polyethylene-plastic (accessed on 7 November 2020).
- Scheirs, J.; Priddy, D. Modern Styrenic Polymers: Polystyrenes and Styrenic Copolymers; John Wiley & Sons: Hoboken, NJ, USA, 2003; p. 3. [Google Scholar]
- XAREC Syndiotactic Polystyrene—Petrochemicals—Idemitsu Kosan Global. Available online: https://www.idemitsu.com/business/ipc/engineering/polystyrene.html (accessed on 7 November 2020).
- Gopi, K.; Balaji, S.; Muthuvelan, B. Isolation, purification and screening of biodegradable polymer PHB producing cyanobacteria from Marine and Fresh Water Resources. Iran. J. Energy Environ. 2014, 5, 94–100. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, L.; An, J.; Chen, B.; Yang, H. Recent strategies for efficient production of polyhydroxyalkanoates by micro-organisms. Lett. Appl. Microbiol. 2016, 62, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Sathiyanarayanan, G.; Bhatia, S.K.; Song, H.S.; Jeon, J.M.; Kim, J.; Lee, Y.K.; Kim, Y.G.; Yang, Y.H. Production and characterization of medium-chain-length polyhydroxyalkanoate copolymer from Arctic psychrotrophic bacterium Pseudomonas sp. PAMC 28620. Int. J. Biol. Macromol. 2017, 97, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Sarkar, B.; Samantaray, D.P.; Daware, A.; Maity, S.; Pattnaik, S.; Bhattacharjee, S. Bioconversion of fish solid waste into PHB using Bacillus subtilis based submerged fermentation process. Environ. Technol. 2017, 38, 3201–3208. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.R. FTIR analysis of polyhydroxyalkanoates by a locally isolated novel Bacillus sp. AS 3-2 from soil of Kadi region, North Gujarat, India. J. Biochem. Technol. 2012, 3, 380–383. [Google Scholar]
- Fetchko, M.; Kitts, A. A user’s guide to BankIt. In The GenBank Submissions Handbook; National Centre for Biotechnology Information US: Bethesda, MD, USA, 2011. [Google Scholar]
- Snedecor, G.W.; Cochran, W.G. Statistical Methods, 7th ed.; Iowa State University Press: Ames, IA, USA, 1980. [Google Scholar]










| Label 1 | Sample Type | Area in Punjab, Pakistan | Global Positioning System Location 2 | No. of Isolates | PHA Accumulation 3 |
|---|---|---|---|---|---|
| 1 | Compost | Kakrali, Gujrat | 32°51′8.9352″ N 74°4′25.8132″ E | 2 | + in all |
| 2 | Soil | Crop field, Mandi-bahauddin | 32°32′48.1″ N 73°28′00.4″ E | 3 | + in isolates 2a, 2c |
| 3 | Effluent | Akzo Nobel Pakistan, paint industry, Lahore | 31°28′38.9″ N 74°20′32.1″ E | 1 | + in all |
| 4 | Soil | Flying Paper industry, Lahore | 31°31′47.4″ N 74°21′57.3″ E | 1 | + in all |
| 5 | Soil | Toyo Nasic Plastic industry, Kot Abdullah, Gujranwala | 32.1112° N 74.1967° E | 2 | + in 5a, − in 5b |
| 6 | Effluent | Royal industries, Pvt. Ltd. Gujranwala | 32°08′03.5″ N 74°10′01.9″ E | 2 | − In all |
| 7 | Soil | A field in Narowal city | 32°06′28.0″ N 74°52′39.2″ E | 3 | + in 7b, 7c |
| 8 | Soil | Asia poultry farm, Phool Nagar, Dina Nath | 31°12′30.7″ N 73°59′43.2″ E | 3 | + in 8b |
| 9 | Effluent | Universal Food Industries, Lahore | 31°34′58.2″ N 74°18′18.8″ E | 3 | +in 9c |
| 10 | Soil | Corn field, Sargodha | 32°02′58.5″ N 72°38′52.5″ E | 1 | − in all |
| 11 | Soil | Shahtaj Sugar Mills, Mandi-Bahauddin | 32°35′19.8″ N 73°27′17.1″ E | 1 | + in all |
| 12 | Soil | Gabol Chowk, Seetpur, Muzaffargarh | 29°14′50.3″ N 70°50′25.0″ E | 3 | + in all |
| 13 | Compost | Kot Shahan, Gujranwala | 32°13′51.2″ N 74°10′45.5″ E | 2 | + in 13c |
| 14 | Soil | Plastic bag landfill site in Mandi-Bahauddin | 32°34′41.2″ N 73°29′26.4″ E | 2 | + in all |
| 15 | Soil | Indus Rice Mills, Mandii-Bahauddin | 32°35′51.3″ N 73°29′27.7″ E | 2 | − in all |
| 16 | Soil | Engi Plastic Industries, Lahore | 31°19′21.8″ N 74°13′42.4″ E | 2 | + in all |
| 17 | Effluent | Toyo Nasic Plastic industry, Kot Abdullah, Gujranwala | 32.1112° N 74.1967° E | 1 | + in all |
| 18 | Soil | Incineration plant, Wah cantt, Rawalpindi | 33°46′47.8″ N 72°41′05.1″ E | 3 | + in 18a, 18b |
| 19 | Effluent | Sewage stream in Gujranwala | 32°11′58.4″ N 74°10′38.3″ E | 2 | + in all |
| 20 | Soil | Jadeed plastic industry, Pirwadhai, Rawalpindi | 33°38′08.8″ N 73°02′05.2″ E | 2 | + in 20a, − in 20b |
| 21 | Soil | Fazal Paper Mills (Pvt.) Ltd., Okara | 30°52′29.8″ N 73°25′09.6″ E | 2 | + in all |
| 22 | Effluent | Griffon Plastic Industries (Pvt) Ltd., Kot Lakhpat, Lahore | 31°27′02.7″ N 74°20′49.8″ E | 1 | − in all |
| 23 | Soil | Lokhadair Landfill, Lahore | 31°37′37.2″ N 74°25′09.0″ E | 3 | + in all |
| 24 | Soil | Wheat crops in Phalia, Mandi-Bahauddin | 32°25′44.7″ N 73°35′27.5″ E | 2 | − in all |
| 25 | Effluent | Z.A. Food Industries, Faisalabad | 31°26′25.1″ N 73°01′31.0″ E | 3 | + in 25a |
| 26 | Effluent | Campbell Flour Mills, Attock | 33°45′30.7″ N 72°21′43.6″ E | 3 | − in all |
| 27 | Soil | Gujrat Road, Phalia, Mandi-Bahauddin | 32°26′08.8” N 73°36′59.3” E | 2 | + in all |
| 28 | Soil | Agbro Poultry Breeding Farms, Jajja-Dhok, Rawalpindi | 33°19′42.2″ N 73°02′14.0″ E | 1 | − in all |
| 29 | Compost | Near Narowal railway station, Narowal | 32°06′02.2″ N 74°51′38.3″ E | 1 | + in all |
| 30 | Soil | Margalla Packages Industry Plastic Bags Manufacturers, Rawalpindi | 33°37′00.1″ N 73°03′09.2″ E | 3 | + in all |
| 31 | Effluent | Century Paper and Board Mills Ltd., Lahore | 31°30′10.3″ N 74°19′30.6″ E | 3 | + in 31a, 31c |
| 32 | Effluent | Sewage stream, Narowal | 32°06′29.9″ N 74°51′43.1″ E | 3 | − in all |
| 33 | Soil | Prime Tanning Industries (Pvt.) Ltd., Sheikhupura | 31°46′53.5″ N 74°15′31.7″ E | 2 | + in 33a |
| 34 | Compost | Alipur, Sheikhupura | 31°43′36.0″ N 74°13′38.8″ E | 3 | + in 34a, 34c |
| 35 | Soil | MashaAllah Poultry Farm, Chak Beli Khan, Rawalpindi | 33°16′21.1″ N 72°54′34.7″ E | 2 | − in all |
| 36 | Effluent | Quick Food Industries Mon Salwa, Lahore | 31°17′04.0″ N 74°10′42.2″ E | 3 | + in 36a, 36b |
| 37 | Effluent | Hassan marbles industry, Jehlum | 32°55′30.9″ N 73°43′09.5″ E | 2 | − in all |
| 38 | Soil | Lucky Plastic Industries (Pvt) Ltd., Raiwind road, Lahore | 31°16′26.3″ N 74°05′47.2″ E | 3 | + in all |
| 39 | Soil | Noon Sugar Mills Ltd., Bhalwal Road, Sargodha | 32°16′44.4″ N 72°55′02.1″ E | 1 | + in all |
| 40 | Soil | Sewage stream in Kasur | 31°06′41.1″ N 74°28′26.4″ E | 1 | + in all |
| 41 | Compost | Sargodha | 32°03′55.6″ N 72°37′58.4″ E | 2 | + in all |
| 42 | Effluent | Askari Cement Ltd., Wah | 33°49′01.3″ N 72°43′25.5″ E | 2 | − in all |
| 43 | Effluent | Jauharabad Sugar Mills Limited, Khushab | 32°18′21.7″ N 72°16′28.5″ E | 1 | + in all |
| 44 | Soil | Wheat field, Sargodha | 32°05′38.2″ N 72°42′49.3″ E | 1 | − in all |
| 45 | Soil | Andrew Paints, Islamabad | 33°32′35.6″ N 73°08′21.3″ E | 2 | + in 45a, − in 45b |
| 46 | Soil | Arsam Pulp and Paper Industries Limited, Sheikhupura | 31°41′15.8″ N 74°02′26.1″ E | 3 | + in 46b, 46c |
| 47 | Soil | Amir Food Industry, Faisalabad | 31°26′53.7″ N 73°05′50.8″ E | 2 | + in all |
| 48 | Soil | Hero Plastic Industries, Lahore | 31°36′27.1″ N 74°21′14.7″ E | 1 | + in all |
| 49 | Compost | Sheikhupura | 31°43′46.5″ N 74°00′33.7″ E | 1 | + in all |
| 50 | Soil | Capital Chemical Industries, Rawalpindi | 33°30′47.9″ N 73°13′52.7″ E | 2 | + in 50a, − in 50b |
| 51 | Effluent | Pakistan Paint Factory, Rawalpindi | 33°38′50.0″ N 73°03′37.1″ E | 2 | + in all |
| 52 | Soil | Dandot Cement Factory, Jehlum | 32°38′39.4″ N 72°58′34.7″ E | 3 | + in 52b |
| 53 | Effluent | Dump site, Sheikhupura | 31°43′47.5″ N 74°00′02.0″ E | 2 | + in all |
| 54 | Soil | Kamalia Sugar Mills, Toba Tek Singh | 30°45′59.7″ N 72°34′49.5″ E | 3 | + in 54a |
| 55 | Soil | Oval Ground, Government College University, Lahore | 31°34′22.2″ N 74°18′24.8″ E | 1 | − in all |
| 56 | Effluent | Anarkali Bazar, Lahore | 31°34′04.7″ N 74°18′16.5″ E | 2 | − in all |
| 57 | Soil | GCU Girls Hostel, Lahore | 31°34′08.1″ N 74°18′16.6″ E | 1 | + in all |
| 58 | Soil | Tariq Plastic Industry, Gujranwala | 32°12′12.7″ N 74°10′22.9″ E | 1 | - in all |
| 59 | Effluent | Expert Advertising and Packaging Ltd., Lahore | 31°26′29.0″ N 74°19′05.2″ E | 2 | + in 59a, − in 59b |
| 60 | Soil | Gujranwala | 32°06′37.7″ N 74°12′35.7″ E | 3 | + in 60c |
| 61 | Soil | Pioneer Cement Ltd., Lahore | 31°31′29.6″ N 74°19′27.4″ E | 2 | − in all |
| 62 | Soil | Qazi Town, Gujranwala | 32°06′41.2″ N 74°12′02.0″ E | 2 | + in 62a, − in 62b |
| 63 | Soil | Glow Paints Factory, Rawalpindi | 33°37′22.5″ N 73°05′46.1″ E | 2 | − in all |
| 64 | Soil | Corn Field, Gujranwala | 32°08′02.0″ N 74°13′30.7″ E | 2 | − in all |
| 65 | Soil | Al-Aziz Plastic Industry, Gujranwala | 32°11′37.9″ N 74°09′38.5″ E | 2 | + in 65a, − in 65b |
| Sample Label (No. of Isolates with +PHA Production) | Isolate | Average PHA Production of the Isolate (%PHA) | Sample Label (No. of Isolates with +PHA Production) | Isolate | Average PHA Production of the Isolate (%PHA) |
|---|---|---|---|---|---|
| 1 (2) | 1a | 19.93 ± 0.25 | 33 (1) | 33a | 38.22 ± 0.24 |
| 1b | 16.15 ± 0.19 | 34 (2) | 34a | 17.25 ± 0.13 | |
| 34c | 12.02 ± 0.28 | ||||
| 2 (2) | 2a | 35.66 ± 0.16 | 36 (2) | 36a | 25.22 ± 0.22 |
| 36b | 21.36 ± 0.15 | ||||
| 2c | 29.27 ± 0.48 | 38 (3) | 38a | 31.25 ± 0.36 | |
| 38b | 26.12 ± 0.23 | ||||
| 38c | 29.55 ± 0.37 | ||||
| 3 (1) | 3a | 43.76 ± 0.18 | 39 (1) | 39a | 36.25 ± 0.51 |
| 4 (1) | 4a | 37.59 ± 0.22 | 40 (1) | 40a | 45.23 ± 0.23 |
| 5 (1) | 5a | 41.2 ± 0.18 | 41 (2) | 41a | 15.96 ± 0.29 |
| 41b | 17.69 ± 0.35 | ||||
| 7 (1) | 7a | 15.22 ± 0.28 | 43 (1) | 43a | 34.51 ± 0.21 |
| 8 (1) | 8b | 52.78 ± 0.23 | 45 (1) | 45a | 38.22 ± 0.45 |
| 9 (1) | 9c | 22.98 ± 0.50 | 46 (2) | 46b | 30.55 ± 0.31 |
| 46c | 28.63 ± 0.26 | ||||
| 11 (1) | 11a | 39.68 ± 0.21 | 47 (2) | 47a | 39.21 ± 0.38 |
| 47b | 35.25 ± 0.48 | ||||
| 12 (3) | 12a | 37.00 ± 0.15 | 48 (1) | 48a | 53.24 ± 0.15 |
| 12b | 8.87 ± 0.17 | ||||
| 12c | 2.09 ± 0.18 | ||||
| 13 (1) | 13c | 19.37 ± 0.39 | 49 (1) | 49a | 21.03 ± 0.14 |
| 14 (2) | 14a | 43.75 ± 0.35 | 50 (1) | 50a | 15.96 ± 0.23 |
| 14b | 16.79 ± 0.46 | ||||
| 16 (2) | 16a | 69.72 ± 0.17 | 51 (2) | 51a | 11.23 ± 0.53 |
| 16b | 55.98 ± 0.19 | 51b | 13.24 ± 0.27 | ||
| 17 (1) | 17a | 24.33 ± 0.21 | 52 (1) | 52b | 8.03 ± 0.19 |
| 18 (2) | 18a | 8.64 ± 0.34 | 53 (2) | 53a | 36.66 ± 0.16 |
| 18b | 38.97 ± 0.27 | 53b | 31.26 ± 0.22 | ||
| 19 (2) | 19a | 11.56 ± 0.42 | 54 (1) | 54a | 17.36 ± 0.18 |
| 19b | 27.67 ± 0.18 | ||||
| 20 (1) | 20a | 35.92 ± 0.31 | 57 (1) | 57a | 20.36 ± 0.35 |
| 21 (2) | 21a | 27.83 ± 0.29 | 59 (1) | 59a | 33.29 ± 0.12 |
| 21b | 28.07 ± 0.23 | ||||
| 23 (3) | 23a | 48.68 ± 0.46 | 60 (1) | 60c | 25.25 ± 0.35 |
| 23b | 26.88 ± 0.28 | ||||
| 23c | 15.68 ± 0.39 | ||||
| 25 (1) | 25a | 12.77 ± 0.38 | 62 (1) | 62a | 29.25 ± 0.11 |
| 27 (2) | 27a | 6.87 ± 0.40 | 65 (1) | 65a | 38.15 ± 0.41 |
| 27b | 12.68 ± 0.20 | ||||
| 29 (1) | 29a | 15.68 ± 0.39 | |||
| 30 (3) | 30a | 30.36 ± 0.27 | |||
| 30b | 25.99 ± 0.19 | ||||
| 30c | 29.11 ± 0.23 | ||||
| 31 (2) | 31a | 19.00 ± 0.11 | |||
| 31c | 18.90 ± 0.28 |
| PCR Primers | Sequencing Primers |
|---|---|
| 27F 5′ (AGA GTT TGA TCM TGG CTC AG) 3′ | 785F 5′ (GGA TTA GAT ACC CTG GTA) 3′ |
| 1492R 5′ (TAC GGY TAC CTT GTT ACG ACT T) 3′ | 907R 5′ (CCG TCA ATT CMT TTR AGT TT) 3′ |
| Parameter | Optimized at | Average PHA Production (%PHA) | Intracellular CDW (g/100 mL) |
|---|---|---|---|
| Incubation time | 24 h | 30.02 ± 0.27 | 0.02 |
| 48 h | 39.64 ± 0.16 | 0.07 | |
| 72 h | 65.39 ± 0.42 | 0.52 | |
| 96 h | 47.56 ± 0.37 | 0.38 | |
| Temperature of incubation | 25 °C | 41.81 ± 0.39 | 0.06 |
| 30 °C | 47.32 ± 0.12 | 0.07 | |
| 37 °C | 78.85 ± 0.23 | 0.59 | |
| 40 °C | 30.41 ± 0.47 | 0.28 | |
| pH of the fermentation medium | 6.0 | 20.14 ± 0.26 | 0.05 |
| 6.5 | 41.70 ± 0.29 | 0.11 | |
| 7.0 | 78.85 ± 0.11 | 0.19 | |
| 7.5 | 32.95 ± 0.33 | 0.07 | |
| 8.0 | 30.09 ± 0.18 | 0.07 | |
| Carbon source | Glucose (standard) | 78.08 ± 0.19 | 0.50 |
| Plastic bag | 68.24 ± 0.27 | 0.19 | |
| Wood chips | 53.15 ± 0.17 | 0.16 | |
| Cardboard cutouts | 51.76 ± 0.48 | 0.15 | |
| Waste shredded polystyrene cups | 43.75 ± 0.30 | 0.17 | |
| Plastic bottle cutouts | 38.19 ± 0.22 | 0.14 |
Sample Availability: Samples of the PHA are available from the authors. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javaid, H.; Nawaz, A.; Riaz, N.; Mukhtar, H.; -Ul-Haq, I.; Shah, K.A.; Khan, H.; Naqvi, S.M.; Shakoor, S.; Rasool, A.; et al. Biosynthesis of Polyhydroxyalkanoates (PHAs) by the Valorization of Biomass and Synthetic Waste. Molecules 2020, 25, 5539. https://doi.org/10.3390/molecules25235539
Javaid H, Nawaz A, Riaz N, Mukhtar H, -Ul-Haq I, Shah KA, Khan H, Naqvi SM, Shakoor S, Rasool A, et al. Biosynthesis of Polyhydroxyalkanoates (PHAs) by the Valorization of Biomass and Synthetic Waste. Molecules. 2020; 25(23):5539. https://doi.org/10.3390/molecules25235539
Chicago/Turabian StyleJavaid, Hadiqa, Ali Nawaz, Naveeda Riaz, Hamid Mukhtar, Ikram -Ul-Haq, Kanita Ahmed Shah, Hooria Khan, Syeda Michelle Naqvi, Sheeba Shakoor, Aamir Rasool, and et al. 2020. "Biosynthesis of Polyhydroxyalkanoates (PHAs) by the Valorization of Biomass and Synthetic Waste" Molecules 25, no. 23: 5539. https://doi.org/10.3390/molecules25235539