Abstract
Farnesyl pyrophosphate synthase (FPS) is a key enzyme that catalyzes the formation of farnesyl pyrophosphate, the main initiator for rubber chain initiation in Hevea brasiliensis Muell. Arg. The transcriptional regulatory mechanisms of the FPS gene still not well understood. Here, a WRKY transcription factor designated HbWRKY27 was obtained by screening the latex cDNA library applied the HbFPS1 promoter as bait. HbWRKY27 interacted with the HbFPS1 promoter was further identified by individual Y1H and EMSA assays. HbWRKY27 belongs to group IIe WRKY subfamily which contains a typical WRKY domain and C-X5-CX23-HXH motif. HbWRKY27 was localized to the nucleus. HbWRKY27 predominantly accumulated in latex. HbWRKY27 was up-regulated in latex by ethrel, salicylic acid, abscisic acid, and methyl jasmonate treatment. Transient expression of HbWRKY27 led to increasing the activity of the HbFPS1 promoter in tobacco plant, suggesting that HbWRKY27 positively regulates the HbFPS1 expression. Taken together, an upstream transcription factor of the key natural rubber biosynthesis gene HbFPS1 was identified and this study will provide novel transcriptional regulatory mechanisms of the FPS gene in Hevea brasiliensis.
Similar content being viewed by others
Introduction
Rubber tree (Hevea brasiliensis Muell. Arg.) is an important rubber-producing plant of Euphorbiaceae1,2. The valuable of rubber tree as a sole commercial source rubber-producing plant led to enormous interest in understanding the natural rubber biosynthesis and regulation in rubber tree. Natural rubber is produced from the latex which is a complex cytoplasmic system of laticifers in the rubber tree3. Generally, natural rubber cis 1, 4-polyisoprene biopolymer, is mainly synthesized by the mevalonate pathway that produces isopentenyl pyrophosphate (IPP) as the precursor and building rubber chain skeleton1. The biosynthetic pathway of natural rubber can be divided into three stages: initiation, polymerization, and termination4. Farnesyl pyrophosphate (FPP) may be the main initiator during natural rubber biosynthesis in rubber tree5,6,7. The concentration of FPP and the ratio of FPP and IPP affect the rubber biosynthetic rate and rubber molecular weight8. Farnesyl pyrophosphate synthase (FPS) catalyzes the consecutive head-to-tail condensations of geranyl pyrophosphate or dimethylallyl diphosphate with two molecules of IPP to form FPP9,10. Thus, FPS should be considered as a crucial enzyme in the natural rubber biosynthesis. The rubber tree FPS genes (named HbFPS1, HbFPS2, and HbFPS3) have been cloned and characterized11. The expression of HbFPS1 exhibits a positive correlation with natural rubber biosynthesis11,12. Recently two MYB transcription factors (HblMYB19 and HblMYB44) are identified to up-regulate the expression of HbFPS113. However, the regulatory mechanisms of the HbFPS1 expression still remain poorly understood. Here, a WRKY transcription factor (designated as HbWRKY27) bound the HbFPS1 promoter and positively regulate HbFPS1 expression, demonstrating that HbWRKY27 might a positive transcription regulator of HbFPS1.
Materials and methods
Plant materials
Rubber tree cultivar CATAS 7-33-97, planted in the experimental plantation of the Chinese Academy of Tropical Agricultural Sciences, was employed to harvest different samples including latex, leaves, flowers, roots, and bark as described previously11. Rubber tree shoots were treated by 0.5% abscisic acid (ABA), 0.2% salicylic acid (SA), 0.07% methyl jasmonate (JA), and 0.5% Ethrel (ET) in accordance with Hao and Wu’ method14. Five groups (10 trees in each group) were employed in each treatment, in which the plant hormone was applied at 3, 6, 9, 12, and 24 h before tapping. The other group was not treated with hormone as control. After the treatments at all time points, latex from all the tested trees were collected. Latex from the same group was mixed together thoroughly. The resulting solution was then divided into five equal volumes for RNA extraction. N. benthamiana seeds were sowed on moist filter paper in a glass garden, and then incubate them in a growth chamber maintained at a relative humidity of 60–70%, a temperature of 28 °C, and 14 h day/10 h night cycle. After a week, the seedlings were potted in soil and placed in a greenhouse maintained at 26–28 °C, a relative humidity of 60–70%, and 14 h day/10 h night cycle. Two months old seedlings were used to test.
Isolation of DNA and RNA
Genomic DNA isolated from young leaves of CATAS 7-33-97 using the Plant Genomic DNA Extraction Kit (TaKaRa, Dalin, China). Isolation of total RNA from different samples was carried out in accordance with the method of Wang et al.15.
Yeast one-hybrid (Y1H) assay
The HbFPS1 promoter fragment (1066 bp) was cloned by PCR with primers (Table 1) using the Genomic DNA as the template in accordance with described method10. Then the HbFPS1 promoter fragment was inserted in bait vector pHIS2.1, generate the pHIS-pHbFPS1 construct. Latex cDNA library was constructed in accordance with the user manual of Matchmaker Gold Yeast One-Hybrid Library Screening System Kit (Clontech, CA, USA). The screening was performed according to the protocol of Matchmaker Gold Y1H System (Clontech, CA, USA). More than 1 × 106 clones were screened and the positive clones were sequenced and analyzed. 35 transformants were obtained and 22 positive colonies were further obtained after re-streaking the primary positive colonies on the same selective medium. These colonies were further analyzed by plasmid rescue followed by sequence analysis.
The interaction between HbWRY27 and the promoter of HbFPS1 was further confirmed by individual Y1H assays. HbWRKY27 was amplified by PCR (see Table 1 for PCR primers) and cloned into the prey vector pGADT7-Rec2 to generate pGADT7-HbWRKY27. pHIS-pHbFPS1 and pGADT7-HbWRKY27 were co-transformed into the yeast Y187 strain. p53-HIS2, pGAD-Rec2-53, as well as pHIS-pHbFPS1 were employed as control. The introduced cells was examined on SD/-Leu-His plates and triple dropout (TDO) plates (SD/-Trp-His-Leu ) supplemented with 80 mM 3-amino-1,2,4-triazole (3-AT) for 5 d at 28 °C.
Electrophoretic mobility shift assay (EMSA)
The full length cDNA of HbWRY27 was cloned by PCR (primers see Table 1). The cDNA fragment was cloned into the vector pET-28a, and then introduced into Escherichia coli strain BL21 to product the HbWRKY27 recombinant proteins according to the user manual (Novagen, Madison, WI, USA). The DNA–protein binding reaction was performed by incubating double-stranded DNA of the HbFPS1 promoter or the mutated promoter with purified recombinant protein at room temperature. The W-box in the HbFPS1 promoter was mutated (changing TTGAC to TTGAA) by single-tube ‘megaprimer’ PCR method16. EMSA was performed with SYBR Green and SYPRO Ruby EMSA stains as described manufacturer’s protocol of EMSA kits (Invitrogen, Carisbad, CA, USA).
Phylogenetic analysis
The homologous protein sequences of HbWRKY27 were obtained from GenBank and phylogenetic analysis was carried out based on the neighbor-joining method using MEGA 5.0 software16.
Subcellular localization
The cDNAs of HbWRKY27 was amplified by PCR with the primers (Table 1) and inserted into the pCAMBIA1302 vector containing the green fluorescent protein (GFP) gene, thereby generating pHbWRKY27-GFP. pHbWRKY27-GFP and pCAMBIA1302 vector were transformed into A. tumefaciens strain GV3101 via electroporation. Then A. tumefaciens harboring pCAMBIA1302 or pHbWRKY27-GFP were transformed into onion epidermis by infiltration as previously described17. Introduced onion epidermal cells was analyzed at 2 days after cultured on MS medium. Fluorescence and 4′, 6′-diamidino-2-phenylindole hydrochloride (DAPI) staining were monitored under a confocal microscope (Leica, Wetzlar, Germany).
Expression analyses of HbWRKY27
Expression of HbWRKY27 was analyzed by real-time qPCRs in accordance with the manufacturer’s instruction of SYBR Premix Taq Kit (TaKaRa, Dalin, China). HbACTIN7 was used as a control gene as described previously17. The relative expression level of HbWRKY27 was calculated using the 2−ΔΔCT method19. Three biological repeats were carried out. Data are presented as mean ± SE (n = 3).
Transient expression assay
The HbFPS1 promoter was cloned by PCR (primers see Table 1) and the HbFPS1 promoter and the mutated promoter fragment was inserted into the pGreenII 0800-LUC vector, generating a reporter construct pGreenII-pHbFPS1-LUC and pGreenII-pHbFPS1M-LUC. To generate effector gene, HbWRKY27 was also cloned through PCR primers (Table 1) and inserted into pCAMBIA1301. The generated HbFPS1-LUC HbFPS1M-LUC construct and the effector construct, was introduced into tobacco leaves as previously described20. The dual-luciferase (LUC) assay was performed according to the manufacturer’s protocol of a dual-luciferase reporter assay system (Promega, Fitchburg, WI, USA). More than three biological repeats were carried out. Difference was accepted as significant at P ≤ 0.05.
Results
HbWRY27 interacts with HbFPS1 promoter
The HbFPS1 promoter had been cloned in previous study21. To understand the transcriptional regulatory of HbFPS1, the HbFPS1 promoter was employed as the bait to screen transcription factors that interact with the HbFPS1 promoter from Y1H-based latex cDNA library. Twenty-two colonies were obtained and sequenced. Eight candidates encoding transcription factors were obtained (Supplementary Information Table S1). Among of candidates, one cDNA encoding WRKY transcription factor, named HbWRKY27 according to its homologs in Genbank, was obtained. HbWRKY27 had an open read frame of 1266 bp in length. The molecular mass of the deduced HbWRKY27 protein was 46.9 kDa. HbWRKY27 had a typical WRKY domain and C-X5-CX23-HXH motif (Fig. 1A). HbWRKY27 was classified into group IIe WRKY subfamily (Fig. 1B). A binding site (W-box) for WRKYs in the promoter of HbFPS1 was predicted21. HbWRKY27 interacted with HbFPS1 promoter was further identified by individual Y1H assays (Fig. 2A). To further determine that HbWRKY27 physically bound with the HbFPS1 promoter, EMSA was used to confirm the binding affinity of HbWRKY27 to the HbFPS1 promoter. The recombinant HbWRKY27 protein was obtained by heterologous expressing of HbWRKY27 in E. coli (Fig. 2B). In addition, the W-box in the HbFPS1 promoter was mutated (changing TTGAC to TTGAA) by PCR method. The DNA–protein binding signal was detected with the recombinant HbWRKY27 protein incubated with the HbFPS1 promoter. No binding signal was detected with the mutated HbFPS1 promoter (Fig. 2C). The result of EMSA also displayed HbWRKY27 interacted with HbFPS1 promoter and the TTGAC is necessary for binding of HbWRKY27 protein to the HbFPS1 promoter.
Alignment of the deduced HbWRKY27 protein sequences. (A) WRKY domain (WRKYGQK) and C-X5-CX23-HXH motif of the HbWRKY27. (B) A phylogenetic tree of the HbWRKY27 proteins and other plants group IIe WRKYs was constructed based on the neighbor joining method, including AaWRKY1 (PWA39112), AaWRKY13 (PWA69470), AaWRKY65 (PWA83388), AaWRKY72 (PWA39515), AtWRKY6 (Q9C519), AtWRKY7 (ANM67919), AtWRKY28 (AEE84006), AtWRKY40 (AEE36457), AtWRKY60 (ANM63193), AtWRKY65 (AEE31068), AtWRKY71 (AEE31143), AtWRKY74 (AED93824), CmWRKY10 (AHC54615), CsWRKY6 (AYA73384), GaWRKY107 (AIY62483), GhWRKY60 (AGV75958), NbWRKY17 (AIR74899), OsWRKY14 (DAA05079), OsWRKY16 (DAA05081), OsWRKY28 (Q0DAJ3), OsWRKY32 (DAA05097), OsWRKY49 (DAA05114), OsWRKY68 (DAA05133), PcWRKY4 (AAG35658), VaWRKY71 (AFK27602).
Characterization of the HbWRKY27. (A) Y1H assays of the binding specificity of the HbFPS1 promoter with HbWRKY27. The yeast cells were cultured on a medium lacking leucine, tryptophan, and histidine (SD/–Trp/–Leu/–His) supplemented with 80 mM 3-amino-1,2,4-triazole (3-AT). Panels show yeast serial decimal dilutions. (B) Heterologous expressing of HbWRKY27 in E. coli. 1. Purified HbWRKY27 fusion protein, 2. E. coli cells harboring pET-HbWRKY27 after 3 h of induction, 3. E. coli cells harboring pET-HbWRKY27 not induced, 4. Molecular markers. (C) Analysis of the binding ability of the HbFPS1 promoter with HbWRKY27 was analyzed via electrophoretic mobility shift assay (EMSA). In the left panel, the gel was stained to visualize the DNA with a SYBR green stain. In the right panel, the gel was stained to monitor the proteins with a SYPRO Ruby EMSA stain. Lane 1. The promoter of HbFPS1 DNA (300 ng) only. Lane 2. HbWRKY27 protein (400 ng) with the promoter of HbFPS1 DNA (300 ng). Lane 3. The mutated promoter of HbFPS1 DNA (300 ng) only. Lane 4. HbWRKY27 protein (400 ng) with the mutated promoter of HbFPS1 DNA (300 ng). Lane 5. HbWRKY27 protein (400 ng) only. (D) Subcellular localization of HBWRKY27-GFP fusion protein in onion epidermal cells. GFP was used as a control and DAPI staining as a nuclear marker.
Subcellular localization of HbWRKY27
To elucidate the subcellular localization of HbWRKY27, the green fluorescent protein (GFP) gene was employed as a marker to fuse HbWRKY27 in-frame, generating the HbWRKY27-GFP construct. Compared with the fluorescence was clearly visible in the cytoplasm and nucleus of the cell transformed with 35S-GFP, the fluorescence was restricted to the nucleus of the cell transformed with HbWRKY27-GFP (Fig. 2D), suggesting that HbWRKY27 was a nuclear-localized protein.
Expression profile of HbWRKY27
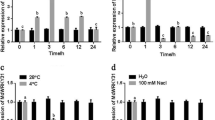
Expression of HbWRKY27 was analyzed by qPCR. The result of qPCR showed that HbWRKY27 predominantly accumulated in latex, but little expression was detected in the leaves, flowers, roots, and barks (Fig. 3A). To investigate HbWRKY27 expression in response to ABA, SA, ET, and JA treatment in latex, qPCR analysis of HbWRKY27 expression were carried out. The expression of HbWRKY27 was up-regulated by ABA, SA, ET, and JA treatment. The expression of HbWRKY27 reached its maximum level after 9 h of SA, ET, and JA treatment, while the expression of HbWRKY27 reached its maximum level after 24 h of ABA treatment (Fig. 3B).
Transcription profiles of HbWRKY27. (A) Expression patterns of HbWRKY27 in rubber tree. Transcript abundances in different tissues are expressed relative to the level in bark. Data are presented as mean ± SE (n = 3). (B) Expression patterns of HbWRKY27 responding to ABA, SA, ET and JA treatment in latex. Transcript abundances in different tissues are expressed relative to the level in control. Data are presented as mean ± SE (n = 3).
HbWRKY27 activates the transcription of HbFPS1
To further study the regulatory relationship of HbWRKY27 and the transcription of HbFPS1, the luciferase (LUC) was employed as a report gene to fuse with the HbFPS1 promoter fragment to generate the pHbFPS1:LUC construct, and the effector p35S-HbWRKY27 was constructed (Fig. 4A). pHbFPS1:LUC was introduced into tobacco leaves along with p35S-GUS or p35S-HbWRKY27. Dual-luciferase assays indicated that HbWRKY27 had significant activation effect on transcription from the HbFPS1 promoter and had no activation effect on transcription from the mutated HbFPS1 promoter (Fig. 4B), indicating that HbWRKY27 could bind the HbFPS1 promoter and activate the HbFPS1 promoter in the transcription level.
Activation of HbFPS1 promoter by HbWRKY27. (A) Schematic drawing of the reporter and effector construct. (B) Effect of HbWRKY27 on the activation of the HbFPS1 promoter. The relative LUC activities (LUC/REN) were normalized to the reference Renilla (REN) luciferase. Error bars indicate SE from five biological replicates (**p < 0.01).
Discussion
FPSs have been identified in a few plants11,22,23,25. FPSs belong to a small multigenic family which encodes at least two different isoforms in plants. The members of the FPS family have a different pattern of expression that vary among different plant species11,22. For example, in Arabidopsis FPS1 is predominantly expressed in roots and inflorescences, whereas FPS2 accumulates preferentially mRNA in inflorescences22. In Ginkgo biloba, the higher GbFPS expression level was detected in roots and leaves23, in which the ginkgolides and bilobalide are synthesized24. In Euphorbia pekinensis, EpFPS had a high transcription level in roots, in which terpenoids are synthesized25. In the rubber tree, HbFPS1 is expressed predominantly in the laticifers and is likely to encode the enzyme involved in natural rubber biosynthesis11. To our knowledge, the transcriptional regulatory mechanisms of FPS gene in plant has not been reported.
WRKY transcription factors, a plant specific transcription factor family, play crucial roles in plant secondary metabolites26,27,28. For example, GaWRKY1 regulates the biosynthesis of gossypol in Gossypium spp29. AaWRKY1 regulates the biosynthesis of artemisinin in Artemisia annua 30. In Vitis vinifera, VviWRKY40 modulates glycosylated monoterpenoid production31. The phylogenetic analyses revealed that HbWRKY27 is highly homologous with MeWRKY27, RoWRKY27, AtWRKY65, and OsWRKY14. OsWRKY14 regulates serotonin production through the up-regulation of the expression of tryptophan synthase gene and tryptophan decarboxylase gene in rice32. The role of other homologs of HbWRKY27 has never been reported. More than 80 WRKY proteins in rubber tree have been identified33. HbWRKY1 is demonstrated to repress the expression of HbSRPP, a natural rubber biosynthesis-related gene, suggesting HbWRKY1 might a negative regulator in natural rubber biosynthesis15. Over-expressing of HbWRKY40 in Arabidopsis increased resistance against Botrytis cinerea34. Except these, the function of few HbWRKYs had been reported. Here, Y1H and EMSA analysis displayed HbWRKY27 bound the HbFPS1 promoter. HbFPS1 is predominantly expressed in latex where natural rubber is synthesized11,12. Intriguingly, HbWRKY27 was also predominantly accumulated in latex, consisting with the expression profile of HbFPS1, suggesting the co-ordinate regulation of natural rubber biosynthesis by both HbWRKY27 and HbFPS1. Moreover transient expression of HbWRKY27 led to increase the activity of the HbFPS1 promoter in vivo, suggesting HbWRKY27 might a positive regulator in natural rubber biosynthesis.
In rubber tree, the natural rubber biosynthesis pathway underlying enzymes have been identified35,36, but the transcriptional regulatory of rubber biosynthesis are poorly understood37,38,39. A few transcription factors except WRKYs had been identified to regulate natural rubber biosynthesis-related gene. For example, HbMADS4 has been identified to negatively regulate HbSRPP expression17, while HbMYC2b positively regulates HbSRPP expression40. HbCZF1 up-regulates 3-hydroxy-3-methyl-glutaryl coenzyme A reductase (HMGR) gene expression41. HblMYB19 and HblMYB44 have been identified to up-regulate the HbFPS1 expression13. HbRZFP1 down-regulates rubber transferase gene (HRT2) expression. The interaction of 14-3-3 protein with HbRZFP1 led to relieve HbRZFP1-mediated HRT2 transcription inhibition42. Even so, the underlying transcriptional regulatory mechanisms of natural rubber biosynthesis are largely unknown. Further investigation of regulatory machinery of natural rubber biosynthesis will be important in manipulating natural rubber metabolism.
Plant hormones have crucial important roles in regulating natural rubber biosynthesis39,43,44. WRKY transcription factors are involved in SA, AB, ET, and JA signaling pathways and plays a vital role in the signal crosstalk of the SA, AB, ET, and JA signaling pathways23,45,46. The promoter of HbWRKY27 had a few cis-acting elements related to hormone responses and HbWRKY27 is simultaneously up-regulated by SA, AB, ET, and JA, suggesting that HbWRKY27 might integrate plant hormones signals and regulates natural rubber biosynthesis. Further investigation should be carried out to study the mechanisms by which HbWRKY27 integrates plant hormones signals and mediates natural rubber biosynthesis.
Conclusion
In the present study, HbWRKY27 was identified to bind the HbFPS1 promoter. HbWRKY27 had significant activation effect on transcription from the HbFPS1 promoter. HbWRK27 might a positive regulator of HbFPS1, which participates in natural rubber biosynthesis.
References
Cornish, K. Similarities and differences in rubber biochemistry among plant species. Phytochemistry 57, 1123–1134 (2001).
van Beilen, J. B. & Poirier, Y. Establishment of new crops for the production of natural rubber. Trends Biotechnol. 25, 522–529 (2007).
d'Auzac, J., Jacob, J.L., & Chrestin, H. The composition of latex from Hevea brasiliensis as laticiferous cytoplasm. In: d'Auzac J, Jacob JL (eds) Physiology of the rubber tree latex. Boca Raton, Florida: CRC Press; pp, 35–42 (1989).
Men, X., Wang, F., Chen, G. Q., Zhang, H. B. & Xian, M. Biosynthesis of natural rubber: current state and perspectives. Int. J. Mol. Sci. 20, 50 (2019).
da Costa, B. M. T., Keasling, J. D. & Cornish, K. Regulation of rubber biosynthetic rate and molecular weight in Hevea brasiliensis by metal cofactor. Biomacromol 6, 279–289 (2005).
Espy, S. C., Keasling, J. D., Castillón, J. & Cornish, K. Initiator-independent and initiator-dependent rubber biosynthesis in Ficus elastica. Arch. Biochem. Biophys. 448, 13–22 (2006).
Xie, W. et al. Initiation of rubber biosynthesis: In vitro comparisons of benzophenone-modified diphosphate analogues in three rubber-producing species. Phytochemistry 69, 2539–2545 (2008).
Cornish, K., Castillón, J. & Scott, D. J. Rubber molecular weight regulation, in vitro, in plant species that produce high and low molecular weights in vivo. Biomacromol 1, 632–641 (2000).
Lange, B. M., Rujan, T., Martin, W. & Croteau, R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 97, 13172–13177 (2000).
Lombard, J. & Moreira, D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol. Biol. Evol. 28, 87–99 (2011).
Guo, D., Li, H. L. & Peng, S. Q. Structure conservation and differential expression of farnesyl diphosphate synthase genes in Euphorbiaceous plants. Int. J. Mol. Sci. 16, 22402–22414 (2015).
Adiwilaga, K. & Kush, A. Cloning and characterization of cDNA encoding farnesyl diphosphate synthase from rubber tree (Hevea brasiliensis). Plant Mol. Biol. 30, 935–946 (1996).
Wang, Y. et al. Transcriptome-wide identification and characterization of MYB transcription factor genes in the laticifer cells of Hevea brasiliensis. Front. Plant Sci. 8, 1974 (2017).
Hao, B. Z. & Wu, J. L. Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann. Bot. 85, 37–43 (2000).
Wang, Y., Guo, D., Li, H. L. & Peng, S. Q. Characterization of HbWRKY1, a WRKY transcription factor from Hevea brasiliensis that negatively regulates HbSRPP. Plant Physiol. Biochem. 71, 283–289 (2013).
Ke, S. H. & Madison, E. L. Rapid and efficient site-directed mutagenesis bysingle-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 25, 3371–3372 (1997).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Li, H. L. et al. HbMADS4, a MADS-box transcription factor from Hevea brasiliensis, negatively regulates HbSRPP. Front. Plant Sci. 7, 170 (2016).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408 (2001).
Hellens, R. G. et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005).
Guo, D., Li, H. L. & Peng, S. Q. Cloning and function analysis of regulatory region for HbFPS1 from Hevea brasiliensis. Trop. Agric. Eng. 34, 33–36 (2010).
Closa, M. et al. The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J. 63, 512–525 (2010).
Wang, P. et al. Cloning and functional analysis of a cDNA encoding Ginkgo biloba farnesyl diphosphate synthase. Mol. Cells 18, 150–156 (2004).
Carrier, D. J., van Beek, T. A., van der Heijden, R. & Verpoort, R. Distribution of ginkgolides and terpenoid biosynthetic activity in G. biloba. Phytochemistry 48, 89–92 (1998).
Cao, X. et al. Molecular characterization and expression analysis of a gene encoding for farnesyl diphosphate synthase from Euphorbia pekinensis Rupr. Mol. Biol. Rep. 39, 1487–1492 (2012).
Outchkourov, N. S. et al. Transcription factor mediated control of anthocyanin biosynthesis in vegetative tissues. Plant Physiol. 176, 1862–1878 (2017).
Eulgem, T. & Somssich, I. E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant. Biol. 10, 366–371 (2007).
Rushton, P. J., Somssich, I. E., Ringler, P. & Shen, Q. J. WRKY transcription factors. Trends Plant Sci. 15, 247–258 (2010).
Xu, Y. H., Wang, J. W., Wang, S., Wang, J. Y. & Chen, X. Y. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 135, 507–515 (2004).
Ma, D. et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161 (2009).
Li, M. X. et al. VviWRKY40, a WRKY transcription factor, regulates glycosylated monoterpenoid production by VviGT14 in grape berry. Genes 11, 485 (2020).
Jin, B. et al. Transcriptome profiling of the spl5 mutant reveals that SPL5 has a negative role in the biosynthesis of serotonin for rice disease resistance. Rice 8, 18 (2015).
Li, H. L., Guo, D., Yang, Z. P., Tang, X. & Peng, S. Q. Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 104, 14–23 (2014).
Yang, J., Wang, Q., Luo, H., He, C. & An, B. HbWRKY40 plays an important role in the regulation of pathogen resistance in Hevea brasiliensis. Plant Cell Rep. 1, 1. https://doi.org/10.1007/s00299-020-02551-x (2020).
Sando, T. et al. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant Hevea brasiliensis. Biosci. Biotechnol. Biochem. 72, 2049–2060 (2008).
Chow, K. S. et al. Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J. Exp. Bot. 58, 242–2440 (2007).
Tang, C. et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2, 16073 (2016).
Yamashita, S. et al. Identification and reconstitution of therubber biosynthetic machinery on rubberparticles from Hevea brasiliensis. eLife 5, e19022 (2016).
Deng, X. et al. Jasmonate signalling in the regulation of rubber biosynthesis in laticifer cells of rubber tree Hevea brasiliensis. J. Exp. Bot. 69, 3559–3571 (2018).
Guo, D., Li, H. L., Wang, Y., Zhu, J. H. & Peng, S. Q. A myelocytomatosis transcription factor from Hevea brasiliensis positively regulates the expression of the small rubber particle protein gene. Ind. Crop. Prod. 133, 90–97 (2019).
Guo, D. et al. Molecular characterization of HbCZF1, a Hevea brasiliensis CCCH-type zinc finger protein that regulates hmg1. Plant Cell Rep. 34, 1569–1578 (2015).
Guo, D. et al. The 14-3-3 protein HbGF14a interacts with a RING zinc finger protein to regulate expression of the rubber transferase gene in Hevea brasiliensis. J. Exp. Bot. 69, 1903–1912 (2018).
Pirrello, J. et al. Transcriptional and post-transcriptional regulation of the jasmonate signalling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. BMC Plant Biol. 14, 341 (2014).
Putranto, R. A. et al. Ethylene response factors are controlled by multiple harvesting stresses in Hevea brasiliensis. PLoS ONE 10, e0123618 (2015).
Xie, Z., Zhang, Z. L., Hanzlik, S., Cook, E. & Shen, Q. J. Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol. Biol. 64, 293–303 (2007).
Fei, X. et al. Patterns of drought response of 38 WRKY transcription factors of Zanthoxylum bungeanum Maxim. Int. J. Mol. Sci. 20, 68 (2018).
Acknowledgements
This study was supported by National Key R&D Program of China (No. 2018YFD1000502) and National Natural Science Foundation of China (No. 31770722).
Author information
Authors and Affiliations
Contributions
S.Q.P. conceived the study. L.Q., H.L.L, D.G., Y.W., and J.H.Z. performed the experiments and carried out the analysis. L.Q., H.L.L, L.Y.Y., and S.Q.P. designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qu, L., Li, HL., Guo, D. et al. HbWRKY27, a group IIe WRKY transcription factor, positively regulates HbFPS1 expression in Hevea brasiliensis. Sci Rep 10, 20639 (2020). https://doi.org/10.1038/s41598-020-77805-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77805-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.