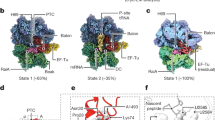
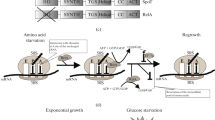
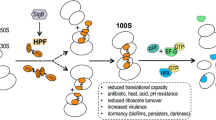
Abstract
In response to stress, eubacteria reduce the level of protein synthesis and either disassemble ribosomes into the 30S and 50S subunits or turn them into translationally inactive 70S and 100S complexes. This helps the cell to solve two principal tasks: (i) to reduce the cost of protein biosynthesis under unfavorable conditions, and (ii) to preserve functional ribosomes for rapid recovery of protein synthesis until favorable conditions are restored. All known genes for ribosome silencing factors and hibernation proteins are located in the operons associated with the response to starvation as one of the stress factors, which helps the cells to coordinate the slowdown of protein synthesis with the overall stress response. It is possible that hibernation systems work as regulators that coordinate the intensity of protein synthesis with the energy state of bacterial cell. Taking into account the limited amount of nutrients in natural conditions and constant pressure of other stress factors, bacterial ribosome should remain most of time in a complex with the silencing/hibernation proteins. Therefore, hibernation is an additional stage between the ribosome recycling and translation initiation, at which the ribosome is maintained in a “preserved” state in the form of separate subunits, non-translating 70S particles, or 100S dimers. The evolution of the ribosome hibernation has occurred within a very long period of time; ribosome hibernation is a conserved mechanism that is essential for maintaining the energy- and resource-consuming process of protein biosynthesis in organisms living in changing environment under stress conditions.



Similar content being viewed by others
Abbreviations
- HPF:
-
hibernation promoting factor
- RaiA:
-
ribosome-associated inhibitor A
- RbfA:
-
ribosome-binding factor A
- RimM:
-
ribosome maturation factor M
- RimP:
-
ribosome maturation factor P
- RMF:
-
ribosome modulation factor
- RsfS:
-
ribosomal silencing factor
REFERENCES
Bergkessel, M., Basta, D. W., and Newman, D. K. (2016) The physiology of growth arrest: uniting molecular and environmental microbiology, Nat. Rev. Microbiol., 14, 549-562, doi: https://doi.org/10.1038/nrmicro.2016.107.
Monod, J. (1947) Kinetics of bacterial growth, Nature, 160, 105-106.
Kolter, R., Siegele, D. A., and Tormo, A. (1993) The stationary phase of the bacterial life cycle, Annu. Rev. Microbiol., 47, 855-874, doi: https://doi.org/10.1146/annurev.mi.47.100193.004231.
Battesti, A., Majdalani, N., and Gottesman, S. (2011) The RpoS-mediated general stress response in Escherichia coli, Annu. Rev. Microbiol., 65, 189-213, doi: https://doi.org/10.1146/annurev-micro-090110-102946.
Gefen, O., Fridman, O., Ronin, I., and Balaban, N. Q. (2014) Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity, Proc. Natl. Acad. Sci. USA, 111, 556-561, doi: https://doi.org/10.1073/pnas.1314114111.
Finkel, S. E. (2006) Long-term survival during stationary phase: evolution and the GASP phenotype, Nat. Rev. Microbiol., 4, 113-120, doi: https://doi.org/10.1038/nrmicro1340.
Williamson, K. S., Richards, L. A., Perez-Osorio, A. C., Pitts, B., McInnerney, K., Stewart, P. S., and Franklin, M. J. (2012) Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population, J. Bacteriol., 194, 2062-2073, doi: https://doi.org/10.1128/JB.00022-12.
Tan, N. C., Cooksley, C. M., Roscioli, E., Drilling, A. J., Douglas, R., Wormald, P. J., and Vreugde, S. (2014) Small-colony variants and phenotype switching of intracellular Staphylococcus aureus in chronic rhinosinusitis, Allergy, 69, 1364-1371, doi: https://doi.org/10.1111/all.12457.
Tuchscherr, L., Medina, E., Hussain, M., Volker, W., Heitmann, V., et al. (2011) Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection, EMBO Mol. Med., 3, 129-141, doi: https://doi.org/10.1002/emmm.201000115.
Ancona, V., Li, W., and Zhao, Y. (2014) Alternative sigma factor RpoN and its modulation protein YhbH are indispensable for Erwinia amylovora virulence, Mol. Plant. Pathol., 15, 58-66, doi: https://doi.org/10.1111/mpp.12065.
Aitken, C. E., Petrov, A., and Puglisi, J. D. (2010) Single ribosome dynamics and the mechanism of translation, Annu. Rev. Biophys., 39, 491-513, doi: https://doi.org/10.1146/annurev.biophys.093008.131427.
Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H., and Noller, H. F. (2001) Crystal structure of the ribosome at 5.5 Å resolution, Science, 292, 883-896, doi: https://doi.org/10.1126/science.1060089.
Ben-Shem, A., Garreau de Loubresse, N., Melnikov, S., Jenner, L., Yusupova, G., and Yusupov, M. (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution, Science, 334, 1524-1529, doi: https://doi.org/10.1126/science.1212642.
Laursen, B. S., Sorensen, H. P., Mortensen, K. K., and Sperling-Petersen, H. U. (2005) Initiation of protein synthesis in bacteria, Microbiol. Mol. Biol. Rev., 69, 101-123, doi: https://doi.org/10.1128/MMBR.69.1.101-123.2005.
Tollerson, R., 2nd, and Ibba, M. (2020) Translational regulation of environmental adaptation in bacteria, J. Biol. Chem., 295, 10434-10445, doi: https://doi.org/10.1074/jbc.REV120.012742.
Korostelev, A. A. (2011) Structural aspects of translation termination on the ribosome, RNA, 17, 1409-1421, doi: https://doi.org/10.1261/rna.2733411.
Pai, R. D., Zhang, W., Schuwirth, B. S., Hirokawa, G., Kaji, H., Kaji, A., and Cate, J. H. (2008) Structural insights into ribosome recycling factor interactions with the 70S ribosome, J. Mol. Biol., 376, 1334-1347, doi: https://doi.org/10.1016/j.jmb.2007.12.048.
Simonetti, A., Marzi, S., Jenner, L., Myasnikov, A., Romby, P., Yusupova, G., Klaholz, B. P., and Yusupov, M. (2009) A structural view of translation initiation in bacteria, Cell. Mol. Life Sci., 66, 423-436.
Nilsson, J., and Nissen, P. (2005) Elongation factors on the ribosome, Curr. Opin. Struc. Biol., 15, 349-354, doi: https://doi.org/10.1016/j.sbi.2005.05.004.
Huter, P., Arenz, S., Bock, L. V., Graf, M., Frister, J. O., et al. (2017) Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P, Mol. Cell, 68, 515-527, doi: https://doi.org/10.1016/j.molcel.2017.10.014.
Golubev, A. A., Validov, S. Z., Usachev, K. S., and Yusupov, M. M. (2019) Elongation factor P: new mechanisms of function and an evolutionary diversity of translation regulation, Mol. Biol., 53, 501-512, doi: https://doi.org/10.1134/S0026893319040034.
Zhou, D., Tanzawa, T., Lin, J., and Gagnon, M. G. (2020) Structural basis for ribosome recycling by RRF and tRNA, Nat. Struct. Mol. Biol., 27, 25-32, doi: https://doi.org/10.1038/s41594-019-0350-7.
Seshadri, A., and Varshney, U. (2006) Mechanism of recycling of post-termination ribosomal complexes in eubacteria: a new role of initiation factor 3, J. Biosciences, 31, 281-289, doi: https://doi.org/10.1007/Bf02703921.
Udagawa, T., Shimizu, Y., and Ueda, T. (2004) Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria, J. Biol. Chem., 279, 8539-8546, doi: https://doi.org/10.1074/jbc.M308784200.
Prossliner, T., Winther, K. S., Sorensen, M. A., and Gerdes, K. (2018) Ribosome hibernation, Annu. Rev. Genet., 52, 321-348, doi: https://doi.org/10.1146/annurev-genet-120215-035130.
McCarthy, B. J. (1960) Variation in bacterial ribosomes, Biochim. Biophys. Acta, 39, 563-564.
Beckert, B., Turk, M., Czech, A., Berninghausen, O., Beckmann, R., et al. (2018) Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1, Nat. Microbiol., 3, 1115-1121, doi: https://doi.org/10.1038/s41564-018-0237-0.
Khusainov, I., Vicens, Q., Ayupov, R., Usachev, K., Myasnikov, A., et al. (2017) Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF, EMBO J., 36, 2073-2087, doi: https://doi.org/10.15252/embj.201696105.
Matzov, D., Aibara, S., Basu, A., Zimmerman, E., Bashan, A., Yap, M. N. F., Amunts, A., and Yonath, A. E. (2017) The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus, Nat. Commun., 8, 723, doi: https://doi.org/10.1038/s41467-017-00753-8.
Polikanov, Y. S., Blaha, G. M., and Steitz, T. A. (2012) How hibernation factors RMF, HPF, and YfiA turn off protein synthesis, Science, 336, 915-918, doi: https://doi.org/10.1126/science.1218538.
Khusainov, I., Fatkhullin, B., Pellegrino, S., Bikmullin, A., Liu, W. T., et al. (2020) Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach, Nat. Commun., 11, 1656, doi: https://doi.org/10.1038/s41467-020-15517-0.
Yamagishi, M., Matsushima, H., Wada, A., Sakagami, M., Fujita, N., and Ishihama, A. (1993) Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase- and growth rate-dependent control, EMBO J., 12, 625-630.
Shimada, T., Yoshida, H., and Ishihama, A. (2013) Involvement of cyclic AMP receptor protein in regulation of the rmf gene encoding the ribosome modulation factor in Escherichia coli, J. Bacteriol., 195, 2212-2219, doi: https://doi.org/10.1128/Jb.02279-12.
Izutsu, K., Wada, A., and Wada, C. (2001) Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp, Genes Cells, 6, 665-676, doi: https://doi.org/10.1046/j.1365-2443.2001.00457.x.
Song, S., and Wood, T. K. (2020) ppGpp ribosome dimerization model for bacterial persister formation and resuscitation, Biochem. Biophys. Res. Commun., 523, 281-286, doi: https://doi.org/10.1016/j.bbrc.2020.01.102.
Sato, A., Watanabe, T., Maki, Y., Ueta, M., Yoshida, H., Ito, Y., Wada, A., and Mishima, M. (2009) Solution structure of the E. coli ribosome hibernation promoting factor HPF: implications for the relationship between structure and function, Biochem. Biophys. Res. Commun., 389, 580-585, doi: https://doi.org/10.1016/j.bbrc.2009.09.022.
Neira, J. L., Giudici, A. M., Hornos, F., Arbe, A., and Rizzuti, B. (2018) The C terminus of the ribosomal-associated protein LrtA is an intrinsically disordered oligomer, Int. J. Mol. Sci., 19, doi: https://doi.org/10.3390/ijms19123902.
Flygaard, R. K., Boegholm, N., Yusupov, M., and Jenner, L. B. (2018) Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism, Nat. Commun., 9, 4179, doi: https://doi.org/10.1038/s41467-018-06724-x.
Boerema, A. P., Aibara, S., Paul, B., Tobiasson, V., Kimanius, D., et al. (2018) Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor, Nat. Plants, 4, 615-615, doi: https://doi.org/10.1038/s41477-018-0203-0.
Usachev, K. S., Fatkhullin, B. F., Klochkova, E. A., Miftakhov, A. K., Golubev, A. A., et al. (2020) Dimerization of long hibernation promoting factor from Staphylococcus aureus: structural analysis and biochemical characterization, J. Struct. Biol., 209, 107408, doi: https://doi.org/10.1016/j.jsb.2019.107408.
Ueta, M., Yoshida, H., Wada, C., Baba, T., Mori, H., and Wada, A. (2005) Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli, Genes Cells, 10, 1103-1112, doi: https://doi.org/10.1111/j.1365-2443.2005.00903.x.
Beckert, B., Abdelshahid, M., Schafer, H., Steinchen, W., Arenz, S., et al. (2017) Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization, EMBO J., 36, 2061-2072, doi: https://doi.org/10.15252/embj.201696189.
Franken, L. E., Oostergetel, G. T., Pijning, T., Puri, P., Arkhipova, V., Boekema, E. J., Poolman, B., and Guskov, A. (2017) A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy, Nat. Commun., 8, 722, doi: https://doi.org/10.1038/s41467-017-00718-x.
McKay, S. L., and Portnoy, D. A. (2015) Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides, Antimicrob. Agents Chemother., 59, 6992-6999, doi: https://doi.org/10.1128/AAC.01532-15.
Yusupov, M. M., and Spirin, A. S. (1986) Are there proteins between the ribosomal subunits? Hot tritium bombardment experiments, FEBS Lett., 197, 229-233, doi: https://doi.org/10.1016/0014-5793(86)80332-5.
Link, A. J., Robison, K., and Church, G. M. (1997) Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12, Electrophoresis, 18, 1259-1313, doi: https://doi.org/10.1002/elps.1150180807.
Agafonov, D. E., Kolb, V. A., Nazimov, I. V., and Spirin, A. S. (1999) A protein residing at the subunit interface of the bacterial ribosome, Proc. Natl. Acad. Sci. USA, 96, 12345-12349, doi: https://doi.org/10.1073/pnas.96.22.12345.
Agafonov, D. E., Kolb, V. A., and Spirin, A. S. (2001) Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage, EMBO Rep., 2, 399-402, doi: https://doi.org/10.1093/embo-reports/kve091.
Di Pietro, F., Brandi, A., Dzeladini, N., Fabbretti, A., Carzaniga, T., Piersimoni, L., Pon, C. L., and Giuliodori, A. M. (2013) Role of the ribosome-associated protein PY in the cold-shock response of Escherichia coli, MicrobiologyOpen, 2, 293-307, doi: https://doi.org/10.1002/mbo3.68.
Hauser, R., Pech, M., Kijek, J., Yamamoto, H., Titz, B., et al. (2012) RsfA (YbeB) proteins are conserved ribosomal silencing factors, PLoS Genet., 8, e1002815, doi: https://doi.org/10.1371/journal.pgen.1002815.
Wanschers, B. F. J., Szklarczyk, R., Pajak, A., van den Brand, M. A. M., Gloerich, J., et al. (2012) C7orf30 specifically associates with the large subunit of the mitochondrial ribosome and is involved in translation, Nucleic Acids Res., 40, 4040-4051, doi: https://doi.org/10.1093/nar/gkr1271.
Jiang, M., Sullivan, S. M., Walker, A. K., Strahler, J. R., Andrews, P. C., and Maddock, J. R. (2007) Identification of novel Escherichia coli ribosome-associated proteins using isobaric tags and multidimensional protein identification techniques, J. Bacteriol., 189, 3434-3444, doi: https://doi.org/10.1128/Jb.00090-07.
Feaga, H. A., Kopylov, M., Kim, J. K., Jovanovic, M., and Dworkin, J. (2020) Ribosome dimerization protects the small subunit, J. Bacteriol., 202, e00009-20, doi: https://doi.org/10.1128/JB.00009-20.
Traub, P., and Nomura, M. (1968) Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins, Proc. Natl. Acad. Sci. USA, 59, 777-784, doi: https://doi.org/10.1073/pnas.59.3.777.
Mulder, A. M., Yoshioka, C., Beck, A. H., Bunner, A. E., Milligan, R. A., et al. (2010) Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit, Science, 330, 673-677, doi: https://doi.org/10.1126/science.1193220.
Nord, S., Bhatt, M. J., Tukenmez, H., Farabaugh, P. J., and Wikstrom, P. M. (2015) Mutations of ribosomal protein S5 suppress a defect in late-30S ribosomal subunit biogenesis caused by lack of the RbfA biogenesis factor, RNA, 21, 1454-1468, doi: https://doi.org/10.1261/rna.051383.115.
Sashital, D. G., Greeman, C. A., Lyumkis, D., Potter, C. S., Carragher, B., and Williamson, J. R. (2014) A combined quantitative mass spectrometry and electron microscopy analysis of ribosomal 30S subunit assembly in E. coli, Elife, 3, e04491, doi: https://doi.org/10.7554/eLife.04491.
Clatterbuck Soper, S. F., Dator, R. P., Limbach, P. A., and Woodson, S. A. (2013) In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates, Mol. Cell, 52, 506-516, doi: https://doi.org/10.1016/j.molcel.2013.09.020.
Wilson, D. N., and Nierhaus, K. H. (2007) The weird and wonderful world of bacterial ribosome regulation, Crit. Rev. Biochem. Mol. Biol., 42, 187-219, doi: https://doi.org/10.1080/10409230701360843.
Huang, Y. J., Swapna, G. V., Rajan, P. K., Ke, H., Xia, B., Shukla, K., Inouye, M., and Montelione, G. T. (2003) Solution NMR structure of ribosome-binding factor A (RbfA), a cold-shock adaptation protein from Escherichia coli, J. Mol. Biol., 327, 521-536, doi: https://doi.org/10.1016/s0022-2836(03)00061-5.
Rubin, S. M., Pelton, J. G., Yokota, H., Kim, R., and Wemmer, D. E. (2003) Solution structure of a putative ribosome binding protein from Mycoplasma pneumoniae and comparison to a distant homolog, J. Struct. Funct. Genom., 4, 235-243, doi: https://doi.org/10.1023/b:jsfg.0000016127.57320.82.
Lovgren, J. M., Bylund, G. O., Srivastava, M. K., Lundberg, L. A., Persson, O. P., Wingsle, G., and Wikstrom, P. M. (2004) The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits, RNA, 10, 1798-1812, doi: https://doi.org/10.1261/rna.7720204.
Leong, V., Kent, M., Jomaa, A., and Ortega, J. (2013) Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement, RNA, 19, 789-802, doi: https://doi.org/10.1261/rna.037523.112.
Grishin, N. V. (2001) KH domain: one motif, two folds, Nucleic Acids Res., 29, 638-643, doi: https://doi.org/10.1093/nar/29.3.638.
Anantharaman, V., and Aravind, L. (2002) The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism, Genome Biol., 3, RESEARCH0061, doi: https://doi.org/10.1186/gb-2002-3-11-research0061.
Guo, Q., Goto, S., Chen, Y., Feng, B., Xu, Y., et al. (2013) Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process, Nucleic Acids Res., 41, 2609-2620, doi: https://doi.org/10.1093/nar/gks1256.
Berti, A. D., Shukla, N., Rottier, A. D., McCrone, J. S., Turner, H. M., et al. (2018) Daptomycin selects for genetic and phenotypic adaptations leading to antibiotic tolerance in MRSA, J. Antimicrob. Chemother., 73, 2030-2033, doi: https://doi.org/10.1093/jac/dky148.
Yoshida, H., and Wada, A. (2014) The 100S ribosome: ribosomal hibernation induced by stress, Wiley Interdiscip. Rev. RNA, 5, 723-732, doi: https://doi.org/10.1002/wrna.1242.
Ueta, M., Wada, C., Daifuku, T., Sako, Y., Bessho, Y., et al. (2013) Conservation of two distinct types of 100S ribosome in bacteria, Genes Cells, 18, 554-574, doi: https://doi.org/10.1111/gtc.12057.
Basu, A., Shields, K. E., and Yap, M. F. (2020) The hibernating 100S complex is a target of ribosome-recycling factor and elongation factor G in Staphylococcus aureus, J. Biol. Chem., 295, 6053-6063, doi: https://doi.org/10.1074/jbc.RA119.012307.
Funding
The work was supported by the Russian Foundation for Basic Research (project no. 20-34-70021).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare no conflict of interest. This article does not contain studies involving humans or other animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Usachev, K.S., Yusupov, M.M. & Validov, S.Z. Hibernation as a Stage of Ribosome Functioning. Biochemistry Moscow 85, 1434–1442 (2020). https://doi.org/10.1134/S0006297920110115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297920110115