Abstract
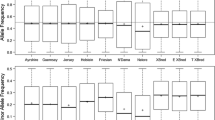
A panel of 200 single nucleotide polymorphisms (SNPs) have been recommended by the International Society for Animal Genetics (ISAG) for use in parentage verification of cattle. While the SNPs included on the ISAG panel are segregating in European Bos taurus and Bos indicus breeds, their applicability in South African (SA) Sanga cattle has never been evaluated. This study, therefore, assessed the usefulness of the ISAG panel in SA Bonsmara (BON) and Drakensberger (DRB) cattle. Genotypes of 185 ISAG SNPs from 64 BON and 97 DRB sire-offspring pairs were available, all of which were validated with 119,375 SNPs. Of the 185 ISAG SNPs, 14 and 18 in the BON and DRB, respectively (9 in common to both breeds), were either monomorphic, exhibited at least one discordance between validated sire-offspring pairs, or had poor call rate or clustering issue. The mean minor allele frequency of the 185 ISAG SNPs was 0.331 in the BON and 0.359 in the DRB. The combined probability of parentage exclusion (PE) was the same (99.46%) for both breeds, while the probability of identity varied from 1.61 × 10−48 (BON) to 1.11 × 10−54 (DRB). Fifteen (23.4%) and 32 (33%) of the already validated sire-offspring pairs for the BON and DRB, respectively, were determined by the ISAG panel to be false-negatives based on a threshold of having at least two discordant SNPs. In comparison to sire discovery using the 119,375 SNPs, sire discovery using only the ISAG panel identified correctly 44 (out of 64 identified using the 119,375 SNPs) unique sire-offspring BON pairs and 62 (out of 97 identified using the 119,375 SNPs) unique sire-offspring DRB when all sires were masked. Five BON and three DRB offspring had > 1 sire nominated. This study demonstrated that the use of the ISAG panel may result in incorrect exclusions and multiple candidate sires for a given animal. Selection of more informative SNPs is, therefore, necessary in the pursuit of a low-cost and effective SNP panel for indigenous cattle breeds in SA.





Similar content being viewed by others
References
Banos, G., Wiggans, G.R., Powell, R.L., 2001. Impact of paternity errors in cow identification on genetic evaluations and international comparisons. Journal of Dairy Science, 84, 2523-2529.
Berry, D.P., Wall, E. and Pryce, J., 2014. Genetics and genomics of reproductive performance in dairy and beef cattle. Animal, 8(S1), 105-121.
Berry, D.P., McHugh, N., Wall, E., McDermott, K., O’Brien, A.C., 2019. Low-density genotype panel for both parentage verification and discovery in a multi-breed sheep population. Ireland Journal of Agricultural and Food Research, 58, 1-12.
Buchanan, J.W., Woronuk, G.N., Marquess, F.L., Lang, K., James, S.T., Deobald, H., Welly, B.T., Van Eenennaam, A.L., 2016. Analysis of validated and population specific SNP parentage panels in pedigreed and commercial beef cattle populations. Canadian Journal of Animal Science, 97, 231-240.
Chan, E.K.F., Hawken, R. and Reverter, A., 2008. The combined effect of SNP-marker and phenotype attributes in genome-wide association studies. Animal Genetics, 40, 149-156.
Fernández, M.E., Goszczynski, D.E., Lirón, J.P., Villegas-Castagnasso, E.E., Carino, M.H., Ripoli, M.V., Rogberg-Muñoz, A., Posik, M.D., Peral-García, P. and Giovambattista, G., 2013. Comparison of the effectiveness of microsatellites and SNP panels for genetic identification, traceability and assessment of parentage in an inbred Angus herd. Journal of Genetics and Molecular Biology, 36, 185-191.
Fisher, P.J., Malthus, B., Walker, M.C., Corbett, G., Spelman, R.J., 2009. The number of single nucleotide polymorphisms and on-farm data required for whole-herd parentage testing in dairy cattle herds. Journal of Dairy Science, 92, 369-374.
García-Ruiz, A., Wiggans, G. R. and Ruiz-López, F. J., 2019. Pedigree verification and parentage assignment using genomic information in the Mexican Holstein population. Journal of Dairy Science, 102, 1806-1810.
Georges, M., Charlier, C. and Hayes, B., 2018. Harnessing genomic information for livestock improvement. Nature Reviews Genetics, 20, 135-156.
Hara, K., Watabe, H., Sasazaki, S., Mukai, F. and Mannen, H., 2010. Development of SNP markers for individual identification and parentage test in a Japanese black cattle population. Journal of Animal Science, 81, 152-157.
Hayes, B.J., 2011. Technical note: Efficient parentage assignment and pedigree reconstruction with dense single nucleotide polymorphism data. Journal of Dairy Science, 94, 2114-2117
Heaton, M.P., Harhay, G.P., Bennett, G.L., Stone, R.T., Grosse, W.M., Casas, E., Keele, J.W., Smith, T.P.L., Chitko-McKown, G.C., Laegreid, W.W., 2002. Selection and use of SNP markers for animal identification and paternity analysis in U.S. beef cattle. Mammalian Genome, 13, 272-281
Henderson, C. R., 1973. Sire evaluation and genetic trends. Animal Breeding and Genetics Symposium in Honor of Dr. Jay L. Lush. Champaign, IL., American Society of Animal Science and American Dairy Science Association, 10-41.
ISAG., 2013. ISAG cattle core and additional SNP panel. www.isag.us/Docs/Cattle-SNP-ISAG-core-additional-panel-2013.xlsx
Israel, C. and Weller, J.I., 2000. Effect of misidentification on genetic gain and estimation of breeding values in dairy cattle populations. Journal of Dairy Science, 83 (1), 181-187.
Jamieson, A., Taylor, C.S., 1997. Comparison of three probability formulae for parentage exclusion. Animal Genetics, 28 (6), 397-400.
Judge, M.M., Kearney, J.F., McClure, M.C., Sleator, R.D., Berry, D.P., 2016. Evaluation of developed low-density genotype panels for imputation to higher density in independent dairy and beef cattle populations. Journal of Animal Science 94 (3), 949-962
Junqueira, V.S., Cardoso, F.F., Oliveira, M.M., Sollero, B.P., Silva, F.F. and Lopes, P.S., 2017. Use of molecular markers to improve relationship information in the genetic evaluation of beef cattle tick resistance under pedigree-based models. Journal of Animal Breeding and Genetics, 134, 14-26.
Kios, D., van Marle-Köster, E. and Visser, C., 2012. Application of DNA markers in parentage verification of Boran cattle in Kenya. Tropical Animal Health Production, 44, 471-476
Lyons, R.E., Buttsworth, S., Waine, D. and Kelly, M., 2015. SNP-based parentage in an Australian cattle industry context: Does one size fit all? Proc. Association for Advancement of Animal Breeding and Genetics, 21, 249-252.
McClure, M.C., McCarthy, J., Flynn, P., Weld, R., Keanne, M., O’Connell, K., Mullen, M.P., Waters, S., Kearney, J.F., 2015. SNP selection for nationwide parentage verification in beef and dairy cattle. Proceeding of the International Committee of Animal Recording (ICAR), Technical series, 19, 175-181. Krakow, Poland, June 10-12. www.icar.org/wp-content/uploads/2015/11/ICAR-Technical-Series-19-Krakow-2015-Proceedings.pdf. (Accessed 20 January 2020)
McClure, M.C., McCarthy, J., Flynn, P., McClure, J.C., Dair, E., O’Connell, D.K., Kearney, J.F., 2018. SNP data quality control in a national beef and dairy Cattle system and highly accurate SNP based parentage verification and identification. Frontiers in Genetics, 9, 84.
Nielsen, R. and Signorovitch, J., 2003. Correcting for ascertainment biases when analyzing SNP data: applications to the estimation of linkage disequilibrium. Theorethical Population Biology, 63 (3), 245-255.
R Core Team., 2018. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org
Sanders, K., Bennewitz, J., Kalm E., 2006. Wrong and missing sire information affects genetic gain in the Angeln dairy cattle population. Journal of Dairy Science, 89 (1), 315-321.
Strucken, E.M., Gudex, B., Ferdosi, M.H., Lee, H.K., Song, K.D., Gibson, J.P., Kelly, M., Piper, E.K., Porto-Neto, L.R., Lee, S.H. and Gondro, C., 2014. Performance of different SNP panels for parentage testing in two East Asian cattle breeds. Animal Genetics, 45, 572-575.
Teneva, A., Todorovska, E., Petrović, M.P., Kusza, S., Perriassamy, K., Petrović, V.C., Andrić, D.O., Gadjev, D., 2018. Short tandem repeats (STR) in cattle genomics and breeding. Biotechnology in Animal Husbandry, 34, 127-147.
Van Doormaal, B.J., Kistemaker, G.J. and Johnston, J., 2016. Parentage analysis services for dairy cattle in Canada. Interbull Bulletin, 50, 2-8. Puerto Varas, Chile, October 24-28. www.icar.org/Documents/DNA-WG/Docs/2017-02-20%20Meeting%20Package.pdf. (Accessed 12 October 2019)
Van Eenennaam, A.L., 2016. Use of genetic marker information in beef cattle selection. Proceedings of the Applied Reproductive Strategies in Beef Cattle, 234-241. Des Moines, Iowa, September 7-8.
Visscher, P.M., Woolliams, J.A., Smith, D., Williams, J.L., 2002. Estimation of pedigree errors in the UK dairy population using microsatellite markers and the impact on selection. Journal of Dairy Science, 85, 2368-2375.
Waits, L.P., Luikart, G., Taberlet, P., 2001. Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Molecular Ecology, 10 (1), 249-256.
Weller, J.I., Feldmesser, E., Golik, M., Tager-Cohen, I., Domochovsky, R., Alus, O., Ron, M., 2004. Factors affecting incorrect paternity assignment in the Israeli Holstein population. Journal of Dairy Science, 87 (8), 2627-2640.
Zhao, F., McParland, S., Kearney, F., Du, L. and Berry, D.P. 2015. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genetics Selection Evolution, 47, 49.
Acknowledgments
We wish to thank the South African Bonsmara and Drakensberger Cattle Breeders Societies for providing access to their genotype and pedigree databases available at the Agricultural Research Council (ARC) and South African Studbook.
Funding
The authors would like to acknowledge the funding from the Technology Innovation Agency (TIA) provided to the Beef Genomics Project (BGP). The authors express their gratitude to the projects funded by the Red Meat Research and Development of South Africa (RMRD SA) under the University of Pretoria (UP) that donated with the genotypes. This research project was also financially supported by the ARC, National Research Foundation (NRF), and the UP.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM1
(XLSX 17.6kb)
Rights and permissions
About this article
Cite this article
Sanarana, Y.P., Maiwashe, A., Berry, D.P. et al. Evaluation of the International Society for Animal Genetics bovine single nucleotide polymorphism parentage panel in South African Bonsmara and Drakensberger cattle. Trop Anim Health Prod 53, 32 (2021). https://doi.org/10.1007/s11250-020-02481-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-020-02481-6