Advances in Microtechnology for Improved Cytotoxicity Assessment
- 1BIOMICs Microfluidics Group, Microfluidics Cluster UPV/EHU, University of the Basque Country UPV/EHU, Vitoria-Gasteiz, Spain
- 2Analytical Microsystems & Materials for Lab-on-a-Chip (AMMa-LOAC) Group, Microfluidics Cluster UPV/EHU, Analytical Chemistry Department, University of the Basque Country UPV/EHU, Leioa, Spain
- 3IKERBASQUE, Basque Foundation of Science, Bilbao, Spain
In vitro cytotoxicity testing is essential in the pharmaceutical and environmental industry to study the effects of potential harmful compounds for human health. Classical assays present several disadvantages: they are commonly based on live-death labelling, are highly time consuming and/or require skilled personnel to be performed. The current trend is to reduce the number of required cells and the time during the analysis, while increasing the screening capability and the accuracy and sensitivity of the assays, aiming single cell resolution. Microfabrication and surface engineering are enabling novel approaches for cytotoxicity assessment, offering high sensitivity and the possibility of automation in order to minimize user intervention. This review aims to overview the different microtechnology approaches available in this field, focusing on the novel developments for high-throughput, dynamic and real time screening of cytotoxic compounds.
Introduction
In the last decades, remarkable advancements have been made in the field of microtechnology to improve analytical processes in biology, through miniaturization, for biosensing DNA (Bulyk et al., 1999; Zhang et al., 2010) and protein arrays (He et al., 2008; Ramachandran et al., 2008; Lopez-Alonso et al., 2013; Gonzalez-Pujana et al., 2019), on-chip electrophoresis (Fritzsche et al., 2010; Ou, et al., 2019), microimmunoassays (Riahi et al., 2016; Hu et al., 2017), microfluidic cell sorting (Shields et al., 2015; Vaidyanathan et al., 2018) and for cellular membrane modelling (Hirano-Iwata et al., 2010; Strulson and Maurer, 2011; Galvez et al., 2020), among others (Beebe, et al., 2002; Sackmann, et al., 2014). In fact, microtechnology enables the precise control of the topography and the surface chemistry, leading to engineered platforms for the study of cellular processes or biosensing and, at the same time, bringing advantages such as time saving, reduced costs and working space, automation of the processes, increased sensitivity and reduced volumes of the required reagents (Wurm et al., 2010; Azuaje-Hualde et al., 2017).
Cytotoxicity assays are crucial in basic research, material science, environmental analysis, pharmaceutical industry and also, in the development of anticancer therapies, where the understanding of the resistance of cancer cells to new therapies is of major importance. Fluorescent and colorimetric assays are the most widely used methods for cytotoxicity assessment (Stoddart, 2011). There are many fluorescent dyes in the market for the measurement of cell viability, whose working principle varies from dye exclusion assays (penetrating the cellular membrane of death cells, staining them), DNA condensation-based assays (which emit fluorescence when they bind nucleic acid molecules) and assays monitoring a metabolic function (Ramirez et al., 2010; Stoddart, 2011). Fluorescent dyes are popular because they are sensitive and provide an easy readout by fluorescence or optical microscopy. Besides, flow cytometry is a well stablished technique for the assessment of cellular toxicity, which combined with some of the previously mentioned alive/dead staining, enables the performance of highly sensitive analysis of cellular viability with single cell resolution (Adan et al., 2017). Since fluorescence gives a semi-quantitative analogic signal and many fluorescent dyes are compatible with single cell analysis, they are often combined with flow cytometry to obtain absolute and real quantification of alive and dead cells within a population. Besides the noteworthy resolution and efficacy of flow cytometry, it requires trained personnel to perform it, as well as staining procedures and manual handling, which take time and increase the possibility of human errors. Furthermore, flow cytometry does not permit real-time monitoring (Gelles and Chipuk, 2016). Therefore, miniaturized systems that reduce manual handling procedures and facilitate fast and easy measurements of cellular viability are promising alternatives to fight the current drawbacks in cytotoxicity assessment, such as the manual intervention, use of labels and the lack of real time information or monitoring.
Nowadays, the large variety of microfabrication techniques leads to the manufacturing of microsystems of many different natures. All the miniaturized platforms for cytotoxicity testing can be usually combined with traditional cellular viability assays, resulting on assays with the same working principle of the traditional staining protocols for the signalling of alive and dead cells, but with improved performance. This is due to the improvements that microtechnology brings in terms of real-time monitoring capability, increased sensitivity, reduced times for the cytotoxicity assessment, decreased volumes of the required reagents and thus, produces a lower cost per analysis. Also, thanks to their small dimensions, it opens the door to multiplexed and multisampling analysis, increasing the yield in the performed protocols (Wang et al., 2007; Sugiura et al., 2008). Furthermore, alternative working principles for the assessment of cytotoxicity are now available due to the sensitivity gained by working at the microscale.
This review aims to summarize the state of the art of the microtechnology used in cytotoxicity tests, and to discuss the novelty that microfabrication techniques and materials bring to this field of research. In this work, we aspire to build a bridge between microfabrication, material science and cytotoxicity, highlighting the advantages brought to the field. In order to do that, the most common microfabrication techniques, microsystems and materials for cytotoxicity will be first explained. Then, the different types of micro analytical platforms existing for cytotoxicity assessment will be comprehensively reviewed (Figure 1).
Microfabrication Techniques Used in Cytotoxicity
Microfabrication techniques were originally developed for the microelectronics industry, specifically for the manufacturing of microelectromechanical systems (MEMs), making possible the integration of sensors to develop fully functional microdevices. Due to the high precision and resolution of the features achieved by these techniques, they soon became useful for the control of tissues and cells at the microscale, becoming attractive for biosensing, leading to the development of BioMEMs (Eribol et al., 2016). The biocompatibility of the materials used in bio-applications is crucial, e.g., the microdevices, in order to avoid any cellular toxic response. Therefore, micromachining for BioMEMs requires adaptations of the fabrication processes and/or the materials used for device fabrication in order to guarantee cellular viability (Fischer et al., 2011).
At the microscale, any small perturbation on the flow or a high density of adhered cells can cause huge stress to them, resulting on exhaustion of nutrients or even migration, which can be misunderstood as a cellular response to the toxic (Wang et al., 2007). These phenomena can be avoided through microfabrication; on one hand, inducing physical cell adhesion control by patterning small microwells on polymeric materials (Vajrala et al., 2019; Zhang et al., 2019) or on C- (Lee et al., 2006) and on U-shaped (Wang et al., 2007) microsieves in the culturing chambers. On the other hand, cell adhesion can be chemically controlled with micropatterns of proteins (Liu et al., 2013; Hamon et al., 2016) or hydrophilic-hydrophobic sites (Nath et al., 2004; Zhang et al., 2007), modulating cell adhesion by enhancing cell affinity for specific areas, keeping them comfortably located. These methods control cell adhesion density and homogeneity of the pattern. Moreover, in the case of C-shaped microsieves, the flow within the chamber is also slowed down, reducing shear stress in cells (Wang et al., 2007), making the device more appropriate for cell culture.
Sotf-Lithography
Photolithography, a lithographic process that uses optical methods for the printing of features, is the most used technique to make integrated circuits, traditionally on rigid surfaces (Levinson, 2005). In particular, the development of soft-lithography, which derives from photolithography and serves to replicate structures from rigid molds -like the ones made by photolithography- on elastomeric or flexible materials, made a huge breakthrough in the manufacturing of BioMEMs. It opened a range of possibilities to use microstructured elastomeric materials, more suitable to keep cellular comfort and to be closer to the mimicking of cellular environment, (Xia and Whitesides, 1998).
The most common material in soft-lithography is polydimethylsiloxane (PDMS), which allows for the fabrication of devices by fast molding techniques, featuring resolutions down to nanometer scale. Also, due to the deformability of this elastomeric material, it facilitates leak-proof fluidic connections as well as integration of valves and other fluidic components. It can be covalently bonded to another PDMS piece or glass surface by a simple plasma oxidation step, resulting on the generation of leaking free, well-sealed devices. Moreover, PDMS is biocompatible, transparent (Bélanger and Marois, 2001) and non fluorescent (Piruska et al., 2005) therefore, suitable for cell applications.
Due to the high resolution of photolithography, the location of cells can be accurately controlled, and the pattern can be precisely transferred to PDMS. The cell positioning capability of PDMS molds has been demonstrated and used for cytotoxicity assessment using U-shaped microsieves, with minimum separation of 8 μm, patterned within culturing chambers (Figures 2A). These features induced low flow regions and a good distribution of cells along the chamber to obtain cellular monolayers with adequate microenvironment (Wang et al., 2007). In the mentioned work, the percentage of cellular death in response to five toxins (digitonin, saponin, CoCl2, NiCl2 and acrolein) was obtained when using Live/Death staining thanks to an adequate cell positioning. Similar idea was used by Mo et al., who combined a micropillar array for uniform cell positioning with a precisely controlled, low flow-rate gradient to reduce the shear stress to human induced pluripotent stem cells (HiPSC) derived neurospheres, and obtain reliable responses against riluzole drug (Mo et al., 2020).
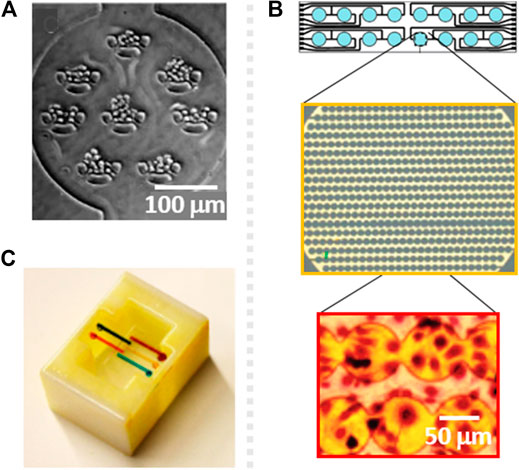
FIGURE 2. (A) Schematic of a cytotoxicity testing microsystem with U-shaped sieves for cell adhesion control, fabricated by soft photolithography. Reproduced from (Wang et al., 2007) with permission from The Royal Society of Chemistry. (B) Design of an electrochemical impedance surface microelectrode array and zoomed picture of the electrodes (yellow) and the cells cultured on top (red), fabricated by electrode patterning. Reprinted with permission from (Xing et al., 2005). (C) 3D printed, reversibly pluggable microfluidic for toxicity screening. Reprinted with permission from (Ding et al., 2015) Copyright © 2015 American Chemical Society.
Soft-lithography is the most popular technique to fabricate gradient generators in microdevices. They generate a gradient in the composition of a solution, like, in this case, the toxic compound to be tested. Thanks to this PDMS gradient generating devices, several works have demonstrated that High-Throughput (HTP) toxicity analysis of specific compounds can be simultaneously carried out, at different concentrations, on chip, for cytotoxicity assessment (Hosokawa et al., 2011; McCormick et al., 2017; Li et al., 2018)
PDMS has been the most commonly used material for the fabrication of prototypes during the last decade but it presents some drawbacks such as absortion of small molecules and high gas permeability, thus there is a demand to fabricate microfluidic devices made of other polymers. As an example, Nguyen et al. reported a hybrid polymeric microfluidic device for cytotoxicity testing, made of poly(methyl methacrylate) with integrated polyethylene terephthalate (Nguyen et al., 2019). The hybrid device was fabricated by milling machine, and was successfully used for cytotoxicity assessment, showing more reliable results than conventional PDMS devices, due to the poly(methyl methacrylate) natural impermeability to small molecules.
Electrode Patterning
Electrode patterning has been one of the most used microfabrication techniques for the monitoring of cytotoxicity responses from the emergence of microtechnology in this field, and it is usually based on additive (metal deposition, insulation), semi-additive (photolithography) and subtractive (etching) processes to fabricate the whole functional electric circuit. The integration or deposition of electrodes on PDMS is challenging (Lim et al., 2006; Guo et al., 2012), so rigid materials such as glass or silicon are commonly used as substrates for electrode patterning, which enables the adequate adhesion of the electrodes on the substrate.
For instance, there are a family of cytotoxicity tests based on gold microelectrode arrays named as xCELLigence Real Time Cell Analyzer (RTCA), which perform continuous monitoring of cytotoxicity by electrical impedance (Ke et al., 2011). This technology relies on the covering of 70–80% of the surface of a well in a 96 well plate with an array of microelectrodes, being this whole area able to measure impedance under a constant voltage (Figures 2B). A variation on the impedance measurements can be attributed to the natural proliferation of cells, to cell detachment and/or to changes in the cytoskeleton-surface interaction due to the response of the cells to external stimuli such as the presence of a toxic compound.
In another work, gold Interdigitated Electrode Structures were fabricated on glass by lithographic techniques, sealed with a PDMS upper part to create a cell culture chamber (Ceriotti et al., 2007). According to the results, the system based on Interdigitated Electrode Structures detected and quantified impedance changes caused by the modulation of cellular adhesion in response to sodium arsenite, like in previously mentioned microelectrode arrays.
Furthermore, due to the resolution of photolithography, surface gold microsensors can be patterned even for the positioning of single cells. For instance Asphahani et al. (2012), used single cell surface electrodes with a diameter of 30 µm to dynamically measure the adhesion of single cells to the electrode substrate in the presence of a hydrogen peroxide (H2O2), sodium arsenate (NaAsO2) or disodium hydrogen arsenate (Na2HAsO4). This system requires the precise position of single cells on the electrodes, but the avoidance of cell adhesion out of the electrodes is crucial too, to help decreasing the noise that can affect the measurement of impedance of single cells. To address this challenge, the authors functionalized the gold electrodes with RGD peptide to induce ligand-mediated natural cell adhesion. As just mentioned, electrode patterning can be complemented with surface modification techniques to modify the gold electrode surface and provide specific capabilities to the sensors. For instance, Nery and co-workers integrated a microarray of gold sensors on glass through common Printed Circuit Board technology. Then, seven different selective membranes were deposited on the sensors, resulting on four ion selective electrodes for K+, Na+, H+, and Ca2+, and three partially selective membrane electrodes (amine-selective, cation-selective and anion- selective), as well as two unmodified gold electrodes for the sensing of red-ox reactions (Nery et al., 2014). The manufacturing and modification of these electrodes lead to the continuous monitoring of cellular death through potentiometric flow-through sensing, correlating changes in the media of the analytes with cellular cytotoxicity.
On the other hand, electrobiofabrication implies the use of electrode signals to specifically pattern electroactive polymers on the electrodes, which in comparison to traditional electrode functionalization strategies simplifies the fabrication of the devices, as demonstrated by Shang et al. (2018). Authors functionalized gold microelectrodes with a bio-based redox capacitor film, which enabled the in situ polymerization of chitosan, a redox-active polymer that amplifies the signal from an electron transfer. The system transduced molecular signals into electronic outputs, which allowed the quantification of cell viability of Caco-2 cells in presence of Triton X-100. Results were compared to the ones obtained by a live/dead fluorescent viability kit. The platform showed high sensitivity to measure molecular interactions by chronocoulometry and correlate them with cellular death. Carbon nanotubes are also promising materials for electrode manufacturing due to their high electrical conductivity, chemical stability and good mechanical properties. Aligned carbon nanotubes have been combined with surface electrode patterning techniques to fabricate sensitive floating surface electrodes. These novel electrodes became efficient tools for the electrophysiological monitoring of cells for cytotoxicity assessment (Ta et al., 2014; Ba et al., 2017). According to the authors, the system is in addition reusable, which avoids device-to-device repeatability issues.
3D Printing
3D printing technology was initially developed for rapid prototyping of devices, but soon, due to the improvements provided in precision, resolution, and repeatability, it became a promising technique for industrial production. Usually, traditional bio-MEM fabrication techniques, e.g., electrode patterning and soft lithography (Ho et al., 2015; Capel et al., 2018), require different types of bulky equipment and imply multiple steps and long fabrication times, in order to get the final product. Techniques like soft-lithography are fine for laboratory scale, but when industrial scale is required, simpler fabrication processes such as 3D printing, are on demand. Furthermore, 3D printing offers a range of biocompatible materials that can be printed with good resolution and in a single step, facilitating large-scale production of devices (Hart et al., 2016; Capel et al., 2018). 3D printing technology is still in its infancy when it comes to cytotoxicity assessing microdevices, but some recently developed platforms suggest that it is a promising tool that may facilitate the manufacturing of cytotoxicity testing microdevices in the near future.
For instance, ultra-high resolution Multijet 3D printing uses UV light to polymerize the photoresist, which is deposited in tiny droplets by the printer to obtain highly complex and well-defined features. Sweet et al. used this technique to fabricate a microdevice, which integrated a network of interconnected 3D enclosed microchannels of 750 µm diameter, for the screening of bacterial cells’ resistance (Sweet et al., 2017). Due to the design and translucency of the material, UV spectroscopy based absorbance measurements were used to in situ determine bacterial cytotoxic response to antibiotics.
Some cytotoxicity platforms reported single 3D printed components such as a modular microfluidic cartridge to regulate reagent volumes during the analysis (Ding et al., 2015) (Figures 2C) or a 3D printed gradient generator to create multi-drug combinations (Chen et al., 2018). In both cases, 3D printing was combined with traditional fluorescent and absorbance cytotoxicity assays. Although just a piece of the system was fabricated by 3D printing, the versatility of the platform was increased, facilitating any modification in the design.
Platforms for Cytotoxicity Assessment by Microtechnology
Microfluidics Combined With Traditional Assays
Cytotoxicity assays are typically based on the colorimetric or fluorescent staining of cells for the determination of cell viability or for metabolite detection in order to identify specific events (Ramirez et al., 2010; Stoddart, 2011). These assays are known to be sensitive and effective but, when combined with microtechnology, multiplexing and reduced manual handling and analysis times, among others, become a reality.
Droplet Microfluidics
Droplet microfluidics provides HTP analysis in cell biology by the generation of microscale droplets through immiscible multiphase flows, resulting on picoliter-scale and monodisperse emulsion droplets. This technique leads to the miniaturization of the bioassays, avoiding capillary effects and evaporation, working as individual bio-microreactors. Moreover, the possibility of using this droplets with non-adherent cells is a reality (Mashaghi et al., 2016). The number of cells encapsulated in each droplet can be slightly modulated following Poisson distribution (Mazutis et al., 2013). Moreover, microdroplets are usually compatible with standard screening microscopies and readouts, enabling the combination with traditional colorimetric and fluorometric cytotoxicity assays. As an example of this combination, Du et al. (2013) measured two live/death fluorescent dyes within each droplet containing cells, when exposed to different concentrations of paclitaxel and 5-fluorouracil anticancer drugs. This was also demonstrated in a previous work, in which the process to make the droplets was consciously optimized to get a good Poisson distribution, allocating 25 cells per droplet with high probability (Yu et al., 2010). Cell viability was investigated in the presence of doxorubicin anticancer drug.
Boedicker et al. generated monodispersed aqueous droplets on chip, and tested the viability of bacterial cells in response to different antibiotics (Boedicker et al., 2008). Every droplet had an integrated fluorescent viability indicator. The encapsulation was carried out in a “stochastic confinement” in order to have higher cell density per droplet, looking for a reduced detection time. In this work, cellular viability was determined in response to different concentrations of seven antibiotics. Sarkar and coworkers have extensive expertize in generating droplets on chip, including droplet formation for cell viability assessment used for the development of anticancer therapies (Sarkar et al., 2015; Sarkar et al., 2020). They first reported a microfluidic droplet generator for the encapsulation of single cancer cells and for the monitoring of the individual cells in presence of doxorubicin anticancer drug with live/dead staining. They evaluated the sensitivity of cells to the drug, by monitoring their viability (Sarkar et al., 2015). More recently, they used the same droplet technology to compare immunotherapy with Natural Killer cells with different cell types, and determined that the treatment based on the combination of herceptin and CD16+ Natural Killer cells was more effective for SKOV3 cells than for HER2 (Sarkar et al., 2020).
The HTP formation of droplets has also been reported on a PDMS nanowell array fabricated by soft lithography. Coupled with another PDMS holding layer for the containment of the oil, the droplets were formed when a cell suspension was added through a capillary, creating the droplets with the cells encapsulated (Du et al., 2013). Ethidium bromide and calcein staining were then used to assess cell viability in the presence of 5-fluorouracil, in the droplets, by fluorescence microscopy.
On the other hand, surface engineering techniques are tools that enable de patterning of droplets on surface, at specific locations. Geyer et al. used UV-initiated photografting of a hydrophilic polymer whose hydrophilicity was inverted by UV through a photomask to create superhydrophilic-superhydrophobic micropatterns on nanoporous polymer films, with drastic difference in wettability to spontaneously create an array of separated aqueous microdroplets (Geyer et al., 2011). These patterns were then used by Popova et al. for cytotoxicity testing (Popova et al., 2015). Each of these droplets worked as individual microreactors for cell culturing, and by a sandwiching technique, the doxorubicin anticancer drug was specifically added to the droplets. Using fluorescent viability staining, the alive and dead cells, in the presence of the doxorubicin, were observed in a HTP and pipetting-free way (Figures 3A).
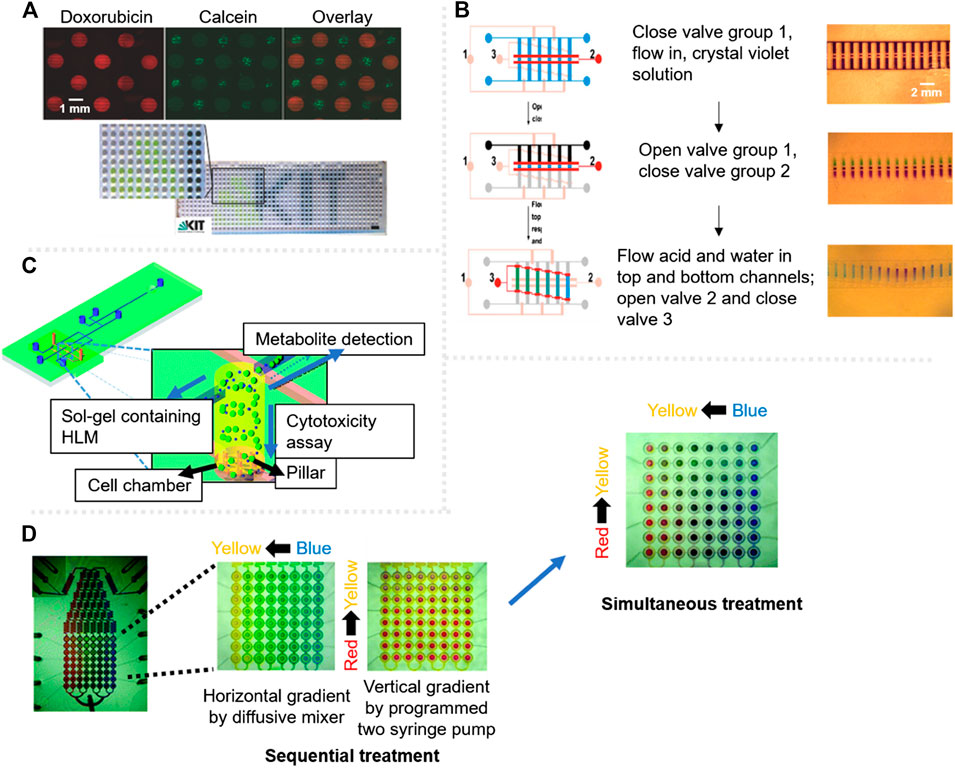
FIGURE 3. Cytotoxicity assessment microsystems with fluorescent label based detection. (A) Droplet based platform showing cells after 18 h in the presence of doxorubicin drug (red) and calcein stained cells for viability test (green); the scale bar in bottom picture is 3 mm. Adapted from (Popova et al., 2015). (B) Schematic of the stepwise gradient operation on chip, signalling valves with colors, red for the open ones and pink for the closed ones. The fluidic channels are fabricated in a ladder shape, each colored channel represents a different solution. Adapted from (Dai et al., 2010). (C) Scheme of a three-layered microdevice for the simultaneous characterisation of drug metabolites and cytotoxicity assessment. Adapted from (Ma et al., 2009) with permission from The Royal Society of Chemistry. (D) Integrated microfluidic device for combinational cytotoxicity assessment by sequential treatment or simultaneous treatment. Adapted from (Kim et al., 2012) with permission from The Royal Society of Chemistry.
Gradient Based Microsystems
The formation of gradients in microfluidic devices has applications in the field of cytotoxicity testing since gradient generators enable the screening of different concentrations of a certain toxic compound at a single step (Dai et al., 2010) (Figures 3B). Ruan et al. (2009) reported a hybrid glass-PDMS microfluidic device with a multi-serpentine based gradient generator made by soft lithography. Cellular viability in response to different concentrations of AsO3 and buthionine sulfoximine chemicals was investigated using acridine orange/ethidium bromide viability assay by fluorescence. When a multi-serpentine based gradient generator is combined with a high-density single-cell array, it results in a microsystem for simultaneous cytotoxicity assessment of numerous concentrations of KCN in this case, with single-cell resolution (Hosokawa et al., 2011). However, the flow-rates commonly used in serpentine gradient generators can be harmful for some cell types such as neurons, so in a recent work, an asymmetric microfluidic device was reported for the formation of drug gradients, which requires lower flows to generate gradients, as mentioned in the section above (Mo et al., 2020).
A multiple dilution method was used in a PDMS microfluidic device to improve the creation of a concentration gradient solution, avoiding distinct parts of the cell experiencing different drug concentrations during the test (Dai et al., 2010). Using Fluorescence Resonance Energy Transfer authors were able to measure cellular death by means of CFP-YFP fluorophores, performing simultaneous cytotoxicity testing of multiple concentrations of etoposide anticancer drug in parallel. The use of hydrogels is another approach to generate gradients in microfluidics. Hydrogels are able to form gradients due to their porous internal structure, which can be interestingly modulated to fulfill specific concentration requirements (Ahadian et al., 2014; Cosson and Lutolf, 2014). As an example, an agarose gel on a microelectrode patterned device allowed the cytotoxic electrochemical monitoring under different concentrations of cisplatin and paclitaxel chemotherapy drugs, which were created through the hydrogel (Tran et al., 2013).
3D printing technology opens the possibility of designing all kind of microfluidic circuitry, including the fabrication of advanced gradients (Sochol et al., 2016). Microsystems capable of generating a gradient of three (nitrofurantoin, tetracycline and trimethoprim) (Sweet et al., 2017) or four (doxorubicin, celecoxib, 5-fluorouracil, and cyclophosphamide) (Chen et al., 2018) toxic compounds allow the monitoring of the cytotoxic response against many combinations of compounds in a single run.
Multiplexing
Miniaturization offers multiplex analysis on chip and it has been reported for many bio-applications, increasing the number of analysis with single cell resolution and so cytotoxicity studies (Kersten et al., 2005; Zhao et al., 2015). Wang and co-workers developed a microfluidic device for multiplex cytotoxicity testing (Wang et al., 2007). The device had several independent cell culturing chambers with U-shaped micro sieves designed to allocate about ten cells per sieve, guaranteeing cell-cell contact, adequate stimulation and comfort. That design enabled the HTP cytotoxicity testing of three cell lines and two concentrations of five toxins (digitonin, saponin, CoCl2, NiCl2 and acrolein) simultaneously.
Droplet microfluidics provides with a different approach to perform simultaneous cell viability assessment, testing different concentrations of doxorubicin and daunorubicin drugs. Based on a hydrophilic/hydrophobic patterning, droplet arrays, working as independent bioreactors, can be individually loaded with the toxic compound of interest or concentrations of the toxic, which leads to HTP and even multiplexing analysis of cytotoxicity using fluorescent live/death assays (Popova et al., 2015; Popova et al., 2017).
Another multiplexing approach relies on the analysis of two different events in parallel, instead of measuring the cellular response to different toxic compounds or concentrations. A microsystem was reported for the characterisation of drug metabolites and for the analysis of their cytotoxic response (Ma et al., 2009). The microsystem had a PDMS bottom layer with three microwells with sol-gel containing encapsulated cells. On top, a quartz layer with a microfluidic network allowed the flow of metabolites (Figures 3C). Due to the multilayer design of the device, the sol-gel acted as a filter to separate drug metabolite detection and cytotoxicity testing, successfully fulfilling both analysis with no cross contamination.
Multifunctional materials deserve special attention for multiplexing. In particular, hydrogels are widely applied as nanocarriers for drug delivery purposes due to their stimuli-responsive behavior. Nanocarriers are easily captured by cells, so, their combination with a sensing functional material could lead to the simultaneous drug release and monitoring of the process (Wu et al., 2010). For instance, Wang and co-workers developed hybrid microspheres made of pH responsive acrylic acid and temperature responsive poly-N-isopropilacrylamide, combined with SiO2 photonic crystal microspheres, which are dielectric structures with an energy band that favors the movement of photons (Wang et al., 2018). Temperature and pH responsiveness of these hybrid and inverse opal hydrogel photonic crystal microspheres, enabled the controllable drug loading/release, whereas due to the photonic crystal component, the cellular viability was dynamically monitored at every point during drug release. Actually, when cells were alive, the microenvironment surrounding them was acidic due to their secretion, green, but when the drug was released and apoptosis was triggered, the pH of the microenvironment was neutralized, changing the color of the spheres to red, which could be observed even by naked eye.
Combinatorial Drug Screening
As mentioned above, cytotoxicity tests are crucial in environmental and pharmacological industries, especially when the toxicity of a compound has to be assessed. Toxicology studies are not simple since the effect of a toxic compound can vary pretty much when in contact with other chemical compounds, sometimes causing an additive effect. However, the combination of more than one chemical or drug causes sometimes synergy (greater effect) or antagonism (lower effect), which may be considered in order to address their toxicity, or their effectiveness in case of therapeutic studies (Foucquier and Guedj, 2015). Therefore, combinatorial-drug-testing can decrease toxicity, reduce the development of drug resistance and even increase the effectiveness of therapies.
Combinatorial tests can be either based on the sequential or simultaneous addition of more than one toxic compound. Considering the first situation, a microsystem based on droplet arrays was reported for sequential toxic combinatorial testing in where the cells in the droplets were sequentially stimulated by two drugs, doxorubicin and daunorubicin (Popova et al., 2017). The microsystem enabled the visualization of cellular viability by fluorescence, combining live/death fluorescent staining. The results showed variations in cell viability when using a single drug or the sequential addition of the two drugs.
Sweet et al. developed a platform for simultaneous cytotoxicity analysis of more than one toxic, generating a gradient with the combination of three different compounds (Sweet et al., 2017). In addition, the platform enabled the identification of the relationship between the toxics. Binary combinations between the toxics correlated well with the results previously published in literature (Bollenbach, 2015). On the other hand, the combination of three toxics identified antagonistic (less effective), synergistic (more effective) and suppressive (least effective) regions during drug screening.
Curved microchannels for fluid mixing driven by Dean flow dynamics -the study of flow in curved channels- were integrated in cytotoxicity screening microsystems to generate gradients and mixing of three compounds, two anticancer drugs (doxorubicin and cisplatin) and the cell media, leading to highly efficient fluid mixing and single-step screening of twelve different combinations (Shen et al., 2020). The twelve combinations were simultaneously investigated for cell viability using a fluorescent live/death assay.
Moreover, a microdevice capable of performing combinatorial screening of two drugs (doxorubicin and mitoxantrone) was reported by Kim et al. (2012), either simultaneously or sequentially. The platform contains 64 individual cell-culture chambers, avoiding the metabolites to be transferred from one chamber to another (Figures 3D). The design of the device allowed the sequential combination of compounds using an accurate gradual valve-opening process, first with one of the toxics, and 24 h after, with the second one. The system showed high screening capability and, with the integrated valve-system, the common drawbacks of existing microdevices on metabolite transference to different cell culturing chambers was sorted out.
Moreover, synergistic effect can be also obtained combining photo-therapy with chemotherapy, so Flont et al. evaluated the effect of doxorubicin drug and nano-encapsulated meso-tetraphenylporphyrin photosensitiser on chip (Flont et al., 2020). Using a microfluidic device mimicking the fluid flows, photodynamic therapy with a photosensitiser and chemotherapy with doxorubicin were sequentially applied to non-malignant and cancer ovarian cells simultaneously, demonstrating the synergistic effect of both therapies, leading to a higher rate of death cancer cells according to MTT viability assay.
Label-Free Microsystems
Common cytotoxicity assays based on enzymatic activity or membrane penetration processes require cell labelling for viability studies. Their widely spread use relies on their elevated effectiveness, reliability and sensitivity, which do not compromise cellular functions such as growth and proliferation. Many efforts are being done to enhance their signal and durability during continuous observation under excitation, e.g., conjugating them with quantum dots to avoid photobleaching over long time exposure to light (Zhao et al., 2009; Zhao et al., 2013). Dye labelling, is time consuming, implies multiple steps and often only detects end points, not allowing continuous monitoring, without showing kinetic or dynamic data. Generally, dyes do not drastically affect essential cellular functions such as growth and proliferation, but there are evidences claiming that cell tracing dyes considerably increase living cell stiffness, deeply increasing the Young’s modulus of the cellular membrane and therefore enhance cellular rigidity and adhesion (Lulevich et al., 2009). Hence, there is a current demand on label-free cytotoxicity monitoring systems for the dynamic analysis of cells upon toxic exposure. Microtechnology brings new approaches to assess cytotoxicity, correlating alternative parameters with cellular death, such as cell adhesion or mechanics, which can be hardly measured without microtechnology.
Microsystems Based on Cell-Substrate Interaction
Cell-substrate interactions are key in most of the vital cellular processes, such as proliferation, differentiation and migration. Regardless of the cell death mechanism, when cells are in the presence of a toxic, undergo morphological changes or disturbances in their membrane, compromising its integrity, which may affect cell adhesion to the substrate (Cho et al., 2008). In the past decades, electrochemical impedance spectroscopy has settled as one of the most popular techniques for the dynamic analysis of cell adhesion to a substrate (Ke et al., 2011), named as electric cell-substrate impedance sensing. The microsystems following this working principle, are based on an array of surface gold electrodes manufactured by lithographic techniques. Cells act as insulating particles, when they adhere to the electrodes, the area in contact with the electrolyte coming from the cell culturing medium is reduced, resulting on an increase on the electric resistance and a decrease in the capacitance values (Hug, 2003). Higher resistance and/or lower capacitance imply a higher density of cells covering the electrode. Based on this technology xCELLigence RTCA emerged for the dynamic monitoring of cellular adhesion on a microarray of gold sensors. In general, when cells are in the presence of a toxic compound, cell death is preceded by a number of morphological and biochemical pathways that at the end lead to death. Impedance based cell adhesion biosensors, enable continuous monitoring of cellular responses against a toxic, such as morphological changes, migration and death (Ke et al., 2011). Using this technology, several studies reported applications on cytotoxicity assessment and showed real-time and dynamic monitoring of cellular responses to different toxic compounds (Xing et al., 2005; Urcan et al., 2010; Irelan et al., 2011; Menotti et al., 2017). Even though electrochemical impedance continuously decreases in the presence of the drug, this value cannot be directly correlated with cellular detachment until signal reaches 0. Electrochemical impedance has been proven sensitive for the monitoring of cellular response to toxics. Nevertheless, until signal does not reach 0 (when there is no cell on the electrodes), due to that sensitivity, the platform may be probably detecting not only cellular detachment, but also changes in morphology caused by the presence of the toxic, and not just detachment or death (Atienza et al., 2005; Xing et al., 2005; Abassi et al., 2009; Susloparova et al., 2015). In order to confirm the correlation between change in impedance signal and quantification of cellular death, xCELLigence RTCA can be coupled with common fluorescent assays for a deeper understanding of the cytotoxic response.
Surface chemistry modification at the micro-scale enables the precise positioning of cells, which following specific adhesion patterns, can lead to new functionalities. In a recent work, microcontact-printing, which creates defined patterns of molecules on surfaces by stamping a micropatterned PDMS, was used to create single cell adhesion dots arrays to address cell viability by optical microscopy (Garcia-Hernando et al., 2020). Cell adhesion spots made by fibronectin protein were designed to allocate single cells, attributing an individual value to each cell. Due to this individuality, the technique enables the counting of adhered cells at every point, so, cellular detachment in the presence of K2CrO4 and DMSO chemical compounds was dynamically monitored for several hours, giving real time and quantitative information of cellular death (Figures 4A).
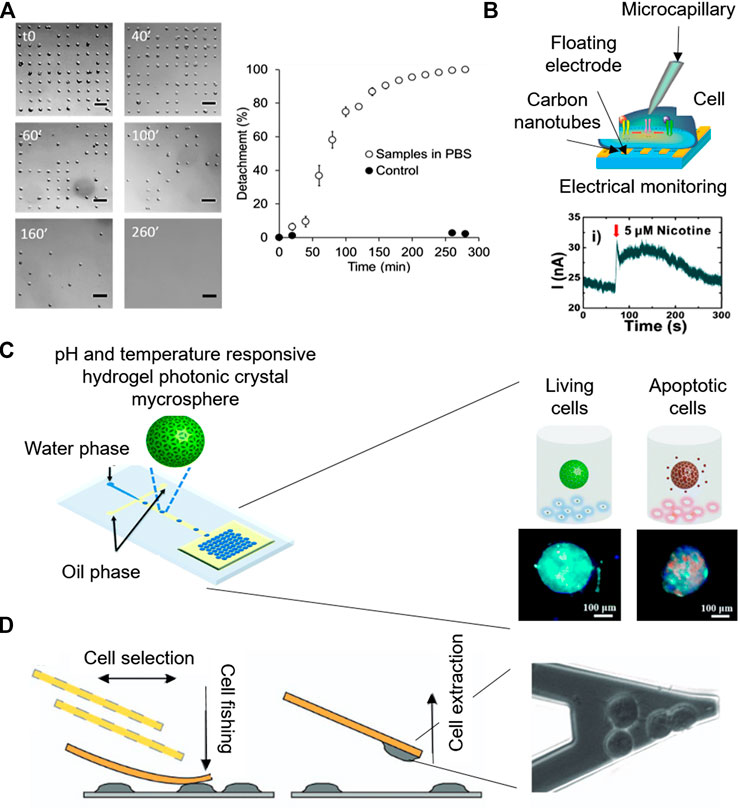
FIGURE 4. Microsystems for label-free cytotoxicity testing. (A) Quantification of cell single-cell adhesion for cytotoxicity assessment by optical microscopy. Adapted from (Garcia-Hernando et al., 2020) Copyright © 2020 American Chemical Society. (B) Working principle of the electrophysiological monitoring of cells for cytotoxicity assessment on aligned carbon nanotubes based gold floating electrodes Adapted with permission from (Ta et al., 2014) Copyright © 2014 American Chemical Society. (C) Schematic of the Photonic crystal microspheres based cytotoxicity platform, showing the control variation under external stimuli for the detection of cell viability. Adapted from (Wang et al., 2018) with permission from The Royal Society of Chemistry. (D) Cytotoxicity monitoring system based on nanomotion sensing of cellular processes. Adapted with permission from (Ruggeri et al., 2017) Copyright © 2017, Springer Nature.
Piezoelectric quartz systems have also been proven useful for live cell biosensing, due to their excellent mass sensing capabilities, leading to the development of mass nanosensors called quartz crystal microbalances (Wang et al., 2011). A quartz crystal microbalances based benchtop platform, comprised of a microplate with a 2 × 2 mm resonant waveguide grating sensor per well was used to monitor cytotoxicity, based on the changes in the dynamic mass redistribution (Farkas et al., 2018). The employed microplate design allowed 384 simultaneous and high-resolution measurements every 3 s, per well. Using the mentioned mass nanosensor, cell death was detected as a negative shift in the dynamic mass redistribution in the presence of toxic compounds. The cytotoxic response is represented in terms of cell size reduction, weakening the adhesion strength and promoting cellular detachment. Therefore, the platform was able to perform non-invasive and continuous measurements for real-time and dynamic monitoring of cytotoxic effects of Roundup Classic, polyethoxylated tallow amine and glyphosate herbicides.
Microsystems Based on the Detection of Specific Molecules
A potentiometric flow-through sensor was reported for label-free cytotoxicity testing. Instead of monitoring cell-substrate adhesion, gold sensors were specifically manufactured and modified to detect multiple parameters that were indirectly correlated with cellular death, such as ions and amines. The potentiometric selective sensing of cations, anions and amines, coupled with partial least squares multivariate statistical analysis, led to non-invasive measurements of cellular viability (Nery et al., 2014).
Quantitative electrophysiological determination of cytotoxic responses in the presence of chemical compounds such as histamine, anti-histamine drugs (chlorphenamine and cetirizine) and acetylcholine receptors (nicotine, daidzein, genistin, and tamoxifen) was carried out using carbon nanotube transistors (Ba et al., 2017; Ta et al., 2014) (Figures 4B). For instance, histamine is a neurotransmitter whose presence in the cellular microenvironment leads to the activation of numerous intracellular electrophysiological pathways, including the opening of the Ca2+ channel gates and the flow of these ions through the plasma membrane. Therefore, due to the quantitative electrochemical signals detected by the floating microsensors, the evaluation of the effect of anti-histamine drugs on non-excitable HeLa cells was carried out (Ba et al., 2017).
Beyond electrochemistry, pH sensing for the label-free and optical assessment of cytotoxicity has also been reported. Photonic crystals are periodic optical nanostructures with very interesting optical properties, due to their atomic energy gap that enables the movements of photons. Photonic crystal microspheres were reported for the colorimetric assessment of cellular viability even by naked eye (Wang et al., 2018). The microspheres were green when cells were alive, because their secretions made their surrounding microenvironment acidic. However, when cells were in presence of the toxic compound and their death was induced, the photonic crystals turned to red because of the neutralization of the cellular microenvironment (Figures 4C).
Mechanics to Monitor Cytotoxicity
The study of cellular mechanics offers a new approach to cytotoxicity assessment, based on a better readout at the cellular level. Cell mechanics results from the overall cellular events, so its study permits to observe the state of the general well-being, but also going to individual cellular events that in conventional assays would be hidden within intrinsic variability.
Due to its precision, atomic force microscopy is a powerful tool for nanomotion sensing in single-cell cytotoxicity tests (Zimmer et al., 2014; Ruggeri et al., 2017).Ruggeri et al. (2017) monitored nanometer-scale movements of cells with the nanomotion sensor, while complementary optical microscopy served to monitor cell attachment, cellular healthiness and micrometer scale movements (Figures 4D). Adding a fluorescent live/dead dye, loss of membrane integrity prior cell death was also detected by nanomotion sensing. Moreover, the cytotoxic responses to amyloids were detected and measured real time.
Single-cell compression method by means of atomic force microscopy can also accurately measure single-cell mechanics in nanotoxicity assessment. Zimmer et al. (2014) used single-cell compression following a protocol previously developed by their group (Lulevich et al., 2006), relying on compressing the cell between a spherical probe and a glass substrate. The cytotoxicity test carried out with this method enabled accurate single-cell mechanic profiles (shrinking, swelling and estimation of Young’s modulus) leading to highly sensitive monitoring of nanotoxicity.
Single Cell Resolution
There is a wide range of options for cytotoxicity assessment of cell populations, including microsystems, bringing new insights in cytotoxicity assessment, such as the HTP capability, multiplexing, gradient formations and label-free dynamic monitoring of cellular viability. However, cell populations are highly heterogeneous, and single cell toxicity analysis could make a difference, identifying cell subpopulations that present higher or lower resistance to a certain dose of a toxic compound. There are two main techniques for the isolation of cells as single cells. On one hand, individual cells can be confined in cavities (Hosokawa et al., 2011), and on the other hand, they can be chemically guided to settle at specific locations by protein patterning techniques (Gonzalez-Pujana et al., 2019), enabling high density of single-cell arrays. Single-cell toxicity testing can provide with quantitative analysis. Fluorescent and colorimetric signals give qualitative data or relatively quantitative values in terms of intensity. However, the identification of single cells as dead or alive provides real numbers and percentages of cellular death, which is actually the basis of the well-stablished flow cytometry technique.
Optical microscopy and label-free monitoring of cytotoxicity is also possible for single-cells. In the recent work from Garcia-Hernando et al., previously mentioned, an array of individual cell adhesion fibronectin islets was used to create single cell arrays for the monitoring of cytotoxicity by optical microscopy (Garcia-Hernando et al., 2020). This system offers two approaches for the assessment of cell viability, depending on the mechanism of action of the toxic compound. The results suggest that when fibronectin-integrin bonding keeps cells attached to the surface, under the effect of some compounds such as K2CrO4 and DMSO cells detach when they die. In this case, the system allows the quantification of cell detachment correlated with cellular death. Contrarily, other chemical compounds such as Hg2SO4 inhibit cell detachment when they die, keeping them dead and individually located on a surface. Adding a colorimetric viability staining, the viability of individual cells can be quantified by optical microscopy.
Agarose platforms, with microcavities for single-cell positioning by gravity, have been reported to measure cellular viability in the presence of nimustine DNA-crosslinker (Li et al., 2016). This microsystem enabled fluorescence microscopy based cellular viability assessment, after staining the cells with a fluorescent live/death assay, giving real percentages of death cells within a population. Hosokawa et al. (2011) also developed a microfluidic device with an array of microcavities to obtain high density single-cell arrays. Those cells were incubated in the presence of KCN and stained with a fluorescent live/death dye, identifying each cell as alive or dead, and giving real quantitative information of cellular viability, with the added value of a gradient generator, leading to a HTP screening of individual cells. In this way, cells were cultured as a population but analyzed as individual cells (Figures 5A). In another work, authors reported a single-cell array cytometry on chip for the analysis of tumor cell apoptosis induced by staurosporine anticancer drug. The microdevice was comprised of 440 mechanical traps of 18 µm width for the hydrodynamic positioning and mechanical trapping of single-cells (Wlodkowic et al., 2009). A fluorescent live/death staining was continuously flowed through the microfluidic device, in a low dose, in order to achieve a continuous labelling without deeply compromising cellular viability, due to the presence of the dye. The system was able to analyze the viability of 250 cells every 1 min, giving real percentages of cellular viability due to the single cell counting. In this particular case, authors obtained dynamic and real-time results of toxic induced cellular death thanks to the continuous loading of a low concentration of fluorescent viability tracker.
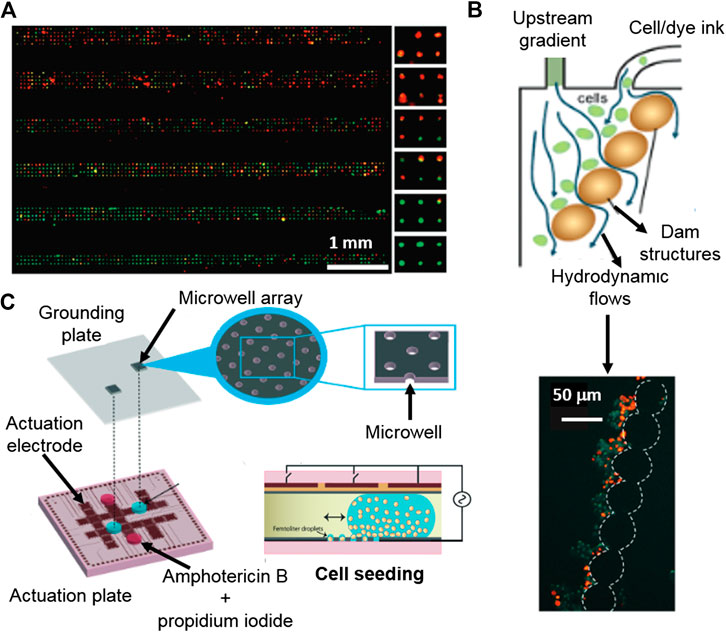
FIGURE 5. Platforms for cytotoxicity assessment of single-cells and non-adherent cells. (A) Gradient generating microfluidic device based on high density single-cell arrays positioned in microcavities for HTP single-cell cytotoxicity testing. Reproduced with permission from (Hosokawa et al., 2011). Copyright © 2011 American Chemical Society. (B) Digital microfluidics device for single-cell toxicity testing based the entrapment of single non-adherent cells. Adapted from (Kumar et al., 2015) with permission from The Royal Society of Chemistry. (C) Microfluidic platform for the cytotoxicity assessment of single-cells isolated by hydrodynamic forces using integrated, dam cell-trapping structures. Adapted with permission from (Zhao et al., 2009) Copyright © 2009 American Chemical Society.
The study of individual cells is important due to the heterogeneity within populations. Electrochemical impedance demonstrates that the dynamic cytotoxic response of individual cells varies from one to others (Asphahani et al., 2012). In this work, the authors reported that not all the single cells gave the same impedance curve, meaning that the cytotoxic response was different from one cell to another, as a clear sign of heterogeneity within the population. Furthermore, the differences in the cytotoxic response were more evident at lower concentrations of the toxic compounds (H2O2, NaAsO2 and Na2HAsO4), giving curves with higher variability. At higher concentrations of toxic compounds, cytotoxic impedance profiles were more similar between single cells, as a sign of high lethality.
Droplet microfluidics can be used for the encapsulation of isolated single-cells by, for instance, coupling droplet formation with an array of microcavities for the deposition of the encapsulated single-cells. An individual cell containing droplet array coupled with a viability fluorescent assay enabled the quantification of individual dead and alive cells, showing also variable cytotoxic responses to antifungal Amphotericin B, within the studied population (Kumar et al., 2015).
Cellular mechanics provide very specific information at the cellular level, offering an alternative readout of the molecular outputs of cellular events. Nanomotion is a promising technique to monitor the cellular well-being, which due to its sensitivity, can monitor cellular movements at the nano-scale in response to α-syn presynaptic protein and SiO2 nanoparticles (Zimmer et al., 2014; Ruggeri et al., 2017).
Non-Adherent Cells
Most of the current existing microsystems for cytotoxicity assessment are meant to be used for adherent cells, since working with them is less tedious and complex than manipulating non-adherent ones. Cytotoxicity assessment of non-adherent cells is mainly performed by flow cytometry, with accurate and reliable results, but again, involves handling of the sample and a temporary gap from the time the experiment is stopped until the time when analysis takes place (Jahan-Tigh et al., 2012). Advances in non-adherent cell viability systems are on demand, since cytotoxicity testing of, for instance, blood cells and circulating tumor cells are crucial in the biomedical industry.
There are not many options available for the entrapment and subsequent analysis of non-adherent cells, being hydrodynamic forces one of them. A microdevice with integrated dam structures for cell-trapping was used to trap cells by hydrodynamic forces, which were applied by an appendant microfluidic channel (Zhao et al., 2009). Once the non-adherent cells were positioned and localized, a live/death fluorescent tracker was used and their intensities were obtained and correlated with the cytotoxic response to the presence of cycloheximide, etoposide and camptothecin antileukemic drugs (Figures 5B).
Droplet microfluidics can be used as tools for the encapsulation and confinement of non-adherent cells. Popova and co-workers demonstrated that an array of droplets made on hydrophilic spots enables the encapsulation and cytotoxicity assessment of T-lymphocytes (Popova et al., 2017). Besides, digital microfluidics permits the confinement of non-adherent individual cells in droplets using an electrowetting-on-dielectric platform. The individually encapsulated cells were positioned as an array of single cells. Then, a stain with cell live/death trackers was used for viability individual cells studies by fluorescence microscopy (Kumar et al., 2015) (Figures 5C).
Floating gold electrode sensors, based on aligned semi-conducing single walled carbon nanotubes, were used for dynamic cytotoxicity monitoring of non-adherent small lung cancer cells in the presence of nicotine and inhibitors of nicotinic acetylcholine receptors, measuring cell’s electrophysiological responses (Ta et al., 2014). When these cells are in a nicotine containing environment, the opening of the Ca2+ channels of the plasma membrane is triggered, causing Ca2+ to flow inside of the cell. This results in a depolarization of the cell, resulting on an increased negative current detected by the electrodes. The sensitivity of the technique enabled the cytotoxicity testing of single-cells.
Automation
Microfabrication enables the integration of several components within the same microsystem, leading to the development of fully integrated devices. They can perform automated processes avoiding handling operations, reducing sample loss and contamination risk, increasing throughput, improving the ease of use and repeatability for, at the end, reduce human errors. Moreover, thanks to automation, remote operation of the microsystems (fluid control and analysis) is possible in case it is required for a particular application (Melin and Quake, 2007).
Many efforts have been made in the automation of microsystems for cytotoxicity testing. Due to the wide use of fluorescent live/dead trackers, automated image processing has been reported in cytotoxicity platforms (Hosokawa et al., 2011; Popova et al., 2017; Li et al., 2018) (Figures 6A). This simplifies data collection, since the platforms can be programmed to take microscopy pictures periodically, for their subsequent analysis, without the need of human supervision.
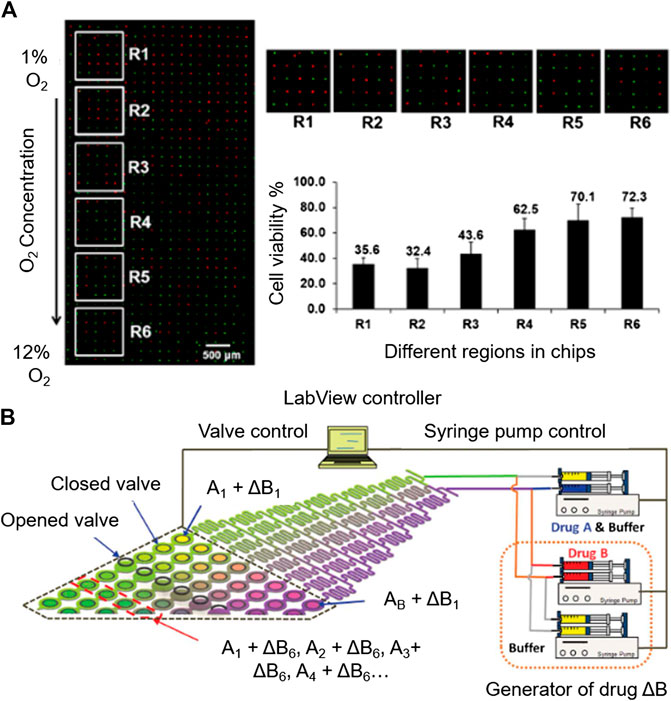
FIGURE 6. Automation potential of microsystems for cytotoxicity assessment. (A) Automated imaging by fluorescence microscopy. Reproduced with permission from (Li et al., 2018). Copyright © 2018 American Chemical Society. (B) Automatable microfluidic system for combinatorial cytotoxicity screening. Reproduced from (Kim et al., 2012) with permission from The Royal Society of Chemistry.
Protocols based on electrochemical monitoring are also compatible with programming during data acquisition, in a dynamic way, far from end-point measurements. xCELLigence RTCA technology is a good example of that, since the manual sample handling is limited to loading the cells into the platform. From this point on, the system continuously measured impedance over time, collecting automatically dynamic data of cells during cytotoxic response (Gurkan et al., 2011; Ke et al., 2011).
Data interpretation can be automated, for instance, when combinatorial toxic screening is performed. The effect of the combined toxics and their concentrations can be automatically uploaded to a combinational library of chemical compounds, facilitating searching for toxic combinations, speeding up the discovery process of unexpectedly harmful toxics or in contrary, finding the most efficient drug combinations for therapy (Ding et al., 2015). On the other hand, Sarkar et al., used algorithms to assess cytotoxicity mediated by Natural Killer cells. A droplet microfluidics generator was used to trap a target cancer cell with a natural killer cell (1:1 ratio), and the interaction between them was accurately studied by time-lapse images, focusing on interaction duration, frequency and cell viability. With the obtained information, algorithms were developed for automatically interpreting the dynamics of natural killer cells as anticancer therapy (Sarkar et al., 2020).
As an example of automated performance is the microfluidic system for sequential combinatorial therapy testing developed by Flont et al. (2020).Going to more complex automation systems, Kim et al., developed a fully programmable microsystem for drug screening (Kim et al., 2012). The platform is comprised of an upstream concentration generation module coupled to an array of 64 individual chambers for cell culturing. The access to each of the cell culture chambers is individually modulated with a valve-array through a pneumatic channel designed to deliver the desired toxic combination to the chambers, controlled by Labview software (Figures 6B). The synchronized valve-system opens the door for a complete automation of the system. This platform simplifies the screening of toxic compound combinations in the investigation of novel effective therapies against cancer.
Nanotoxicology
Nanotoxicology is a subfield of toxicology that focuses on the toxic effects of nano-sized particles, often not caused by the material itself, but by the nano-scale related properties of these particles (Auffan et al., 2009; Catalán et al., 2016; Fadeel, 2019). For instance, the high surface-to-volume ratio of nanoparticles makes them highly reactive when compared to bulk materials (Recordati et al., 2015). Nanoparticles can penetrate cellular membranes, biological barriers and tissues, so they interact with biological systems at the molecular, cellular, organismal and ecosystem levels (Pietroiusti et al., 2018), The increasing interest of using nanoparticles in medicine, opens new possibilities for microtechnology, for instance to mimic whole organs (Organ-on-A-Chip technology), generating devices able to precisely study the effect of nanomaterials in a similar environment to in vivo (Ashammakhi et al., 2020).
Due to the capability of engineered nanomaterials to interact with the cellular membrane—by adsorption, penetration or endocytosis—the study of the interaction between nanomaterials and the cell membrane is important and challenging (Zhang et al., 2012; Churchman et al., 2013). Membrane-on-chip microplatforms emerge for high throughput membrane sensing, giving information of the events happening on a supported cellular membrane. The most used membrane biosensing microtechnology comprises electrochemical sensors fabricated with self-assembled monolayers supported on mercury electrodes that provide information of the interactions or damage on the membrane (Owen et al., 2020; Rashid et al., 2017). Recently, Owen et al. reported an automated, rapid and HTP sensing microfluidic platform for the screening of nanomaterial-biomembrane interactions (Owen et al., 2020). The integrated microfluidic device provided electrochemical information of the interactions of chlorpromazine and citrate coated gold nanomaterial with 1,2-dioleoyl-sn-glycero- 3- phosphocoline membranes, supposing an easy-to-use interface to assess cytotoxicity based on membrane damage (less than 6 min per assay).
Due to the high toxicity and barrier penetration capability of nanoparticles, the studies of their effects on three-dimensional environments, mimicking natural biosystems, enable more reliable approaches to evaluate the physiological effects of nanomaterials (Ashammakhi et al., 2020). For instance, Arends et al. reported a microfluidic device to study the diffusion of nanoparticles through the basal layer, which is key to control the movement of nanoparticles between blood vessels and the surrounding extravascular space, observing a charge dependant accumulation of the species, similar to the one observed in vivo (Arends et al., 2015).
Conclusion
Cytotoxicity assessment is vital in the pharmaceutical and environmental industry for the identification of potentially harmful compounds, as well as for the development of new therapies in medicine. Currently, the most popular techniques are based on labelling cells with fluorescent or colorimetric stains, targeting disrupted membranes of dead cells or specific metabolites. These are very sensitive and effective, but the application of microtechnology in cytotoxicity testing opens the door to more efficient methods that can also be complemented with traditional assays.
Within microtechnology, many techniques have been proven efficient for reliable and sensitive cytotoxicity assessment. It must be highlighted that working at the microscale, HTP approach is possible. Moreover, multiplexed and gradient cytotoxicity assessment systems, capable of collecting higher amounts of data for the correct interpretation of toxicity, become available for general use. Many microdevices for combinatorial toxic screening have been reported, which facilitate research on the identification of potentially toxic compounds, enabling the development of more effective therapies in medicine.
Furthermore, the use of microtechnology for cytotoxicity testing permits the measurement of cellular events such as cell-substrate adhesion, electrophysiology and mechanics of cytotoxicity in a label free manner, avoiding the addition of external labels that even at a low level, could affect cellular functions. Additionally, label-free sensing allows dynamic and continuous monitoring of cells during testing, providing to the user with additional data, e.g., continuous monitoring of cellular well-being, even before death, as well as kinetic responses against toxic compounds.
Besides, cytotoxicity assessment at single-cell level is possible using microtechnology. This enables the study of cytotoxic responses of cell subpopulations within heterogeneous populations, which helps investigating cellular resistance, as well as vulnerabilities of cells to chemical compounds. Single-cell resolution also enables real quantification of cellular viability, which would bring cytotoxicity assessment platforms close to the concept of flow cytometry on a surface. Moreover, the assessment could be automated using programmable microdevices, which facilitates their use, leading to more efficient analyses, reducing human handling and thus, errors.
Nanotoxicology is a recent field within cytotoxicity assessment which is growing fast, because of the emerging interest in nanoparticles for therapy. Reliable information on the effects of engineered nanoparticles on biosystems is crucial, and microtechnology is emerging as a powerful tool for screening nanoparticle—biological system interactions.
Despite the previous advances, microtechnology is still far for being implemented in daily cytotoxicity assessment. Although the presented micro-systems have been successfully implemented for cytotoxicity assessment, their fabrication is still laborious and long, and it sometimes require specific micromachining equipment, complicating the mass production of devices. The use of microplatforms for cytotoxicity testing is very useful for now when it comes to specific cytotoxic measurements or information that cannot be obtained with traditional assays, such as cytotoxicity dynamics, real-time monitoring, gradient formation and multiplexing; or information that can be obtained easily and faster than with traditional tools, such as single cell testing or combinatorial testing. However, microtechnology seems to have already a place in cytotoxicity assessment, and it is lately gaining importance and popularity, considering the increasing number of publications appearing every year with novel approaches. It can be expected that new trends in this field will focus on the improvement of the fabrication of the devices, simplifying processes for potential mass production and looking for suitable materials with properties that permit a good performance of the analyses. Therefore, evidence points out that the future of cytotoxicity assays will be highly conditioned by the developments made in microtechnology.
Author Contributions
MGH: Conceptualization, Ideas, Investigation, Writing - Original Draft, Writing - Review and Editing. FBL and LBD: Conceptualization, Ideas, Resources, Management, Writing - Review and Editing, Supervision, Project administration, Funding acquisition.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors would like to acknowledge the funding support from: University of the Basque Country (PIF16/204), the funding support from Gobierno de España, Ministerio de Economía y Competitividad, with Grant No. BIO2016-80417-P (AEI/FEDER, UE) and Gobierno Vasco under grand IT1271-19.
References
Abassi, Y. A., Xi, B., Zhang, W., Ye, P., Kirstein, S. L., Gaylord, M. R., et al. (2009). Kinetic cell-based morphological screening: prediction of mechanism of compound action and off-target effects. Chem. Biol. 16, 712–723. doi:10.1016/j.chembiol.2009.05.011
Adan, A., Alizada, G., Kiraz, Y., Baran, Y., and Nalbant, A. (2017). Flow cytometry: basic principles and applications. Crit. Rev. Biotechnol. 37, 163–176. doi:10.3109/07388551.2015.1128876
Ahadian, S., Ramón-Azcón, J., Estili, M., Obregón, R., Shiku, H., and Matsue, T. (2014). Facile and rapid generation of 3D chemical gradients within hydrogels for high-throughput drug screening applications. Biosens. Bioelectron. 59, 166–173. doi:10.1016/j.bios.2014.03.031
Arends, F., Sellner, S., Seifert, P., Gerland, U., Rehberg, M., and Lieleg, O. (2015). A microfluidics approach to study the accumulation of molecules at basal lamina interfaces. Lab Chip. 15, 3326–3334. doi:10.1039/c5lc00561b
Ashammakhi, N., Darabi, M. A., Çelebi‐Saltik, B., Tutar, R., Hartel, M. C., Lee, J., et al. (2020). Microphysiological systems: next generation systems for assessing toxicity and therapeutic effects of nanomaterials. Small Methods. 4, 1900589. doi:10.1002/smtd.201900589
Asphahani, F., Thein, M., Wang, K., Wood, D., Wong, S. S., Xu, J., et al. (2012). Real-time characterization of cytotoxicity using single-cell impedance monitoring. Analyst. 137, 3011–3019. doi:10.1039/c2an16079j
Atienza, J. M., Zhu, J., Wang, X., Xu, X., and Abassi, Y. (2005). Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J. Biomol. Screen. 10 (8), 795–805. doi:10.1177/1087057105279635
Auffan, M., Rose, J., Bottero, J., Lowry, G. V., Jolivet, J., and Wiesner, M. R. (2009). Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective. Nat. Nanotechnol. 4, 634–641. doi:10.1038/nnano.2009.242
Azuaje-Hualde, E., García-Hernando, M., Etxebarria-Elezgarai, J., De Pancorbo, M. M., Benito-Lopez, F., and Basabe-Desmonts, L. (2017). Microtechnologies for cell microenvironment control and monitoring. Micromachines. 8, 166. doi:10.3390/mi8060166
Ba, V. A. P., Cho, D., Kim, D., Yoo, H., Ta, V., and Hong, S. (2017). Quantitative electrophysiological monitoring of anti–histamine drug effects on live cells via reusable sensor platforms. Biosens. Bioelectron. 94, 707–713. doi:10.1016/j.bios.2017.03.063
Beebe, D. J., Mensing, G. A., and Walker, G. M. (2002). Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Engineer.,Engineer. 4 (1), 261–286. doi:10.1146/annurev.bioeng.4.112601.125916
Bélanger, M., and Marois, Y. (2001). Hemocompatibility, biocompatibility, inflammatory and in vivo studies of primary reference materials low‐density polyethylene and polydimethylsiloxane: a review. J. Biomed. Mat. Res. 58, 467–477. doi:10.1002/jbm.1043
Boedicker, J. Q., Li, L., Kline, T. R., and Ismagilov, R. F. (2008). Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 8, 1265–1272. doi:10.1039/b804911d
Bollenbach, T. (2015). Antimicrobial interactions: mechanisms and implications for drug discovery and resistance evolution. Curr. Opin. Microbiol. 27, 1–9. doi:10.1016/j.mib.2015.05.008
Bulyk, M. L., Gentalen, E., Lockhart, D. J., and Church, G. M. (1999). Quantifying DNA–protein interactions by double-stranded DNA arrays. Nat. Biotechnol. 17, 573–577. doi:10.1038/9878
Capel, A. J., Rimington, R. P., Lewis, M. P., and Christie, S. D. (2018). 3D printing for chemical, pharmaceutical and biological applications. Nat. Rev. Chem. 2, 422–436. doi:10.1038/s41570-018-0058-y
Catalán, J., Siivola, K. M., Nymark, P., Lindberg, H., Suhonen, S., Järventaus, H., et al. (2016). In vitro and in vivo genotoxic effects of straight versus tangled multi-walled carbon nanotubes. Nanotoxicology. 10, 794–806. doi:10.3109/17435390.2015.1132345
Ceriotti, L., Ponti, J., Colpo, P., Sabbioni, E., and Rossi, F. (2007). Assessment of cytotoxicity by impedance spectroscopy. Biosens. Bioelectron. 22, 3057–3063. doi:10.1016/j.bios.2007.01.004
Chen, X., Chen, H., Wu, D., Chen, Q., Zhou, Z., Zhang, R., et al. (2018). 3D printed microfluidic chip for multiple anticancer drug combinations. Sens. Actuat. B-Chem. 276, 507–516. doi:10.1016/j.snb.2018.08.121
Cho, M., Niles, A., Huang, R., Inglese, J., Austin, C. P., Riss, T., et al. (2008). A bioluminescent cytotoxicity assay for assessment of membrane integrity using a proteolytic biomarker. Toxicol. In Vitro. 22, 1099–1106. doi:10.1016/j.tiv.2008.02.013
Churchman, A. H., Wallace, R., Milne, S. J., Brown, A. P., Brydson, R., and Beales, P. A. (2013). Serum albumin enhances the membrane activity of ZnO nanoparticles. Chem. Commun. 49, 4172–4174. doi:10.1039/c3cc37871c
Cosson, S., and Lutolf, M. (2014). Hydrogel microfluidics for the patterning of pluripotent stem cells. Sci. Rep. 4, 4462. doi:10.1038/srep04462
Dai, W., Zheng, Y., Luo, K. Q., and Wu, H. (2010). A prototypic microfluidic platform generating stepwise concentration gradients for real-time study of cell apoptosis. Biomicrofluidics. 4, 024101. doi:10.1063/1.3398319
Ding, Y., Li, J., Xiao, W., Xiao, K., Lee, J., Bhardwaj, U., et al. (2015). Microfluidic-enabled print-to-screen platform for high-throughput screening of combinatorial chemotherapy. Anal. Chem. 87, 10166–10171. doi:10.1021/acs.analchem.5b00826
Du, G., Pan, J., Zhao, S., Zhu, Y., den Toonder, J. M., and Fang, Q. (2013). Cell-based drug combination screening with a microfluidic droplet array system. Anal. Chem. 85, 6740–6747. doi:10.1021/ac400688f
Eribol, P., Uguz, A., and Ulgen, K. (2016). Screening applications in drug discovery based on microfluidic technology. Biomicrofluidics. 10, 011502. doi:10.1063/1.4940886
Fadeel, B. (2019). The right stuff: on the future of nanotoxicology. Front. Toxicol. 1, 1. doi:10.3389/ftox.2019.00001
Farkas, E., Szekacs, A., Kovacs, B., Olah, M., Horvath, R., and Szekacs, I. (2018). Label-free optical biosensor for real-time monitoring the cytotoxicity of xenobiotics: a proof of principle study on glyphosate. J. Hazard. Mater. 351, 80–89. doi:10.1016/j.jhazmat.2018.02.045
Fischer, R., Steinert, S., Fröber, U., Voges, D., Stubenrauch, M., Hofmann, G., et al. (2011). Cell cultures in microsystems: biocompatibility aspects. Biotechnol. Bioeng. 108, 687–693. doi:10.1002/bit.22951
Flont, M., Jastrzębska, E., and Brzózka, Z. (2020). Synergistic effect of the combination therapy on ovarian cancer cells under microfluidic conditions. Anal. Chim. Acta. 1100, 138–148. doi:10.1016/j.aca.2019.11.047
Foucquier, J., and Guedj, M. (2015). Analysis of drug combinations: current methodological landscape. Pharmaco. Res. Perspect. 3, e00149. doi:10.1002/prp2.149
Fritzsche, S., Hoffmann, P., and Belder, D. (2010). Chip electrophoresis with mass spectrometric detection in record speed. Lab Chip. 10, 1227–1230. doi:10.1039/c000349b
Galvez, J. M. M., Garcia-Hernando, M., Benito-Lopez, F., Basabe-Desmonts, L., and Shnyrova, A. V. (2020). Microfluidic chip with pillar arrays for controlled production and observation of lipid membrane nanotubes. Lab Chip. 20 (15), 2748–2755. doi:10.1039/d0lc00451k
Garcia-Hernando, M., Calatayud-Sanchez, A., Etxebarria-Elezgarai, J., de Pancorbo, M. M., Benito-Lopez, F., and Basabe-Desmonts, L. (2020). Optical single cell resolution cytotoxicity biosensor based on single cell adhesion dot arrays. Anal. Chem. 92 (14), 9658–9665. doi:10.1021/acs.analchem.0c00940
Gelles, J. D., and Chipuk, J. E. (2016). Robust high-throughput kinetic analysis of apoptosis with real-time high-content live-cell imaging. Cell Death & Dis. 7, e2493. doi:10.1038/cddis.2016.332
Geyer, F. L., Ueda, E., Liebel, U., Grau, N., and Levkin, P. A. (2011). Superhydrophobic–superhydrophilic micropatterning: towards genome‐on‐a‐chip cell microarrays. Angew. Chem. Int. Ed. 50, 8424–8427. doi:10.1002/anie.201102545
Gonzalez-Pujana, A., Santos-Vizcaino, E., García-Hernando, M., Hernaez-Estrada, B., de Pancorbo, M. M., Benito-Lopez, F., et al. (2019). Extracellular matrix protein microarray-based biosensor with single cell resolution: integrin profiling and characterization of cell-biomaterial interactions. Sens. Actuat. B- Chem. 299, 126954. doi:10.1016/j.snb.2019.126954
Guo, L., Guvanasen, G. S., Liu, X., Tuthill, C., Nichols, T. R., and DeWeerth, S. P. (2012). A PDMS-based integrated stretchable microelectrode array (isMEA) for neural and muscular surface interfacing. IEEE Trans. Biomed. 7, 1–10. doi:10.1109/TBCAS.2012.2192932
Gurkan, U. A., Anand, T., Tas, H., Elkan, D., Akay, A., Keles, H. O., et al. (2011). Controlled viable release of selectively captured label-free cells in microchannels. Lab Chip. 11, 3979–3989. doi:10.1039/c1lc20487d
Hamon, C., Henriksen‐Lacey, M., La Porta, A., Rosique, M., Langer, J., Scarabelli, L., et al. (2016). Tunable nanoparticle and cell assembly using combined self‐powered microfluidics and microcontact printing. Adv. Funct. Mat. 26, 8053–8061. doi:10.1002/adfm.201602225
Hart, L. R., Li, S., Sturgess, C., Wildman, R., Jones, J. R., and Hayes, W. (2016). 3D printing of biocompatible supramolecular polymers and their composites. ACS Appl. Mat. Interfaces. 8, 3115–3122. doi:10.1021/acsami.5b10471
He, M., Stoevesandt, O., Palmer, E. A., Khan, F., Ericsson, O., and Taussig, M. J. (2008). Printing protein arrays from DNA arrays. Nat. Methods. 5, 175–177. doi:10.1038/nmeth.1178
Hirano-Iwata, A., Oshima, A., Nasu, T., Taira, T., Kimura, Y., and Niwano, M. (2010). Stable lipid bilayers based on micro-and nano-fabrication. Supramol. Chem. 22, 406–412. doi:10.1080/10610278.2010.487564
Ho, C. M. B., Ng, S. H., Li, K. H. H., and Yoon, Y. (2015). 3D printed microfluidics for biological applications. Lab Chip. 15, 3627–3637. doi:10.1039/c5lc00685f
Hosokawa, M., Hayashi, T., Mori, T., Yoshino, T., Nakasono, S., and Matsunaga, T. (2011). Microfluidic device with chemical gradient for single-cell cytotoxicity assays. Anal. Chem. 83, 3648–3654. doi:10.1021/ac2000225
Hu, B., Li, J., Mou, L., Liu, Y., Deng, J., Qian, W., et al. (2017). An automated and portable microfluidic chemiluminescence immunoassay for quantitative detection of biomarkers. Lab Chip. 17, 2225–2234. doi:10.1039/c7lc00249a
Hug, T. S. (2003). Biophysical methods for monitoring cell-substrate interactions in drug discovery. Assay Drug Dev. Techn. 1, 479–488. doi:10.1089/154065803322163795
Irelan, J. T., Wu, M., Morgan, J., Ke, N., Xi, B., Wang, X., et al. (2011). Rapid and quantitative assessment of cell quality, identity, and functionality for cell-based assays using real-time cellular analysis. J. Biomol. Screen. 16, 313–322. doi:10.1177/1087057110397359
Jahan-Tigh, R. R., Ryan, C., Obermoser, G., and Schwarzenberger, K. (2012). Flow cytometry. J. Invest. Dermatol. 132, 1–6. doi:10.1038/jid.2012.282
Ke, N., Wang, X., Xu, X., and Abassi, Y. A. (2011). The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol. Biol. 740, 33–43. doi:10.1007/978-1-61779-108-6_6
Kersten, B., Wanker, E. E., Hoheisel, J. D., and Angenendt, P. (2005). Multiplex approaches in protein microarray technology. Expert. Rev. Proteomic. 2, 499–510. doi:10.1586/14789450.2.4.499
Khalid, N., Kobayashi, I., and Nakajima, M. (2017). Recent lab‐on‐chip developments for novel drug discovery. Wires Syst. Biol. Med. 9, e1381. doi:10.1002/wsbm.1381
Kim, J., Taylor, D., Agrawal, N., Wang, H., Kim, H., Han, A., et al. (2012). A programmable microfluidic cell array for combinatorial drug screening. Lab Chip. 12, 1813–1822. doi:10.1039/c2lc21202a
Kumar, P., Vriens, K., Cornaglia, M., Gijs, M., Kokalj, T., Thevissen, K., et al. (2015). Digital microfluidics for time-resolved cytotoxicity studies on single non-adherent yeast cells. Lab Chip. 15, 1852–1860. doi:10.1039/c4lc01469c
Lee, P. J., Hung, P. J., Rao, V. M., and Lee, L. P. (2006). Nanoliter scale microbioreactor array for quantitative cell biology. Biotechnol. Bioeng. 94, 5–14. doi:10.1002/bit.20745
Li, L., Li, Y., Shao, Z., Luo, G., Ding, M., and Liang, Q. (2018). Simultaneous assay of oxygen-dependent cytotoxicity and genotoxicity of anticancer drugs on an integrated microchip. Anal. Chem. 90, 11899–11907. doi:10.1021/acs.analchem.8b02070
Li, L., Wang, W., Ding, M., Luo, G., and Liang, Q. (2016). Single-cell-arrayed agarose chip for in situ analysis of cytotoxicity and genotoxicity of DNA cross-linking agents. Anal. Chem. 88, 6734–6742. doi:10.1021/acs.analchem.6b01008
Lim, K. S., Chang, W., Koo, Y., and Bashir, R. (2006). Reliable fabrication method of transferable micron scale metal pattern for poly (dimethylsiloxane) metallization. Lab Chip. 6, 578–580. doi:10.1039/b514755g
Liu, W., Li, Y., Wang, T., Li, D., Fang, L., Zhu, S., et al. (2013). Elliptical polymer brush ring array mediated protein patterning and cell adhesion on patterned protein surfaces. ACS Appl. Mat. Interfaces. 5, 12587–12593. doi:10.1021/am403808s
Lopez-Alonso, A., Jose, B., Somers, M., Egan, K., Foley, D. P., Ricco, A. J., et al. (2013). Individual platelet adhesion assay: measuring platelet function and antiplatelet therapies in whole blood via digital quantification of cell adhesion. Anal. Chem. 85, 6497–6504. doi:10.1021/ac401076s
Lulevich, V., Shih, Y., Lo, S. H., and Liu, G. (2009). Cell tracing dyes significantly change single cell mechanics. J. Phys. Chem. B. 113, 6511–6519. doi:10.1021/jp8103358
Lulevich, V., Zink, T., Chen, H., Liu, F., and Liu, G. (2006). Cell mechanics using atomic force microscopy-based single-cell compression. Langmuir. 22, 8151–8155. doi:10.1021/la060561p
Ma, B., Zhang, G., Qin, J., and Lin, B. (2009). Characterization of drug metabolites and cytotoxicity assay simultaneously using an integrated microfluidic device. Lab Chip. 9, 232–238. doi:10.1039/b809117j
Mashaghi, S., Abbaspourrad, A., Weitz, D. A., and van Oijen, A. M. (2016). Droplet microfluidics: a tool for biology, chemistry and nanotechnology. Trac-Trend. Anal. Chem. 82, 118–125. doi:10.1016/j.trac.2016.05.019
Mazutis, L., Gilbert, J., Ung, W. L., Weitz, D. A., Griffiths, A. D., and Heyman, J. A. (2013). Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 8, 870. doi:10.1038/nprot.2013.046
McCormick, S. C., Kriel, F. H., Ivask, A., Tong, Z., Lombi, E., Voelcker, N. H., et al. (2017). The use of microfluidics in cytotoxicity and nanotoxicity experiments. Micromachines. 8, 124. doi:10.3390/mi8040124
Melin, J., and Quake, S. R. (2007). Microfluidic large-scale integration: the evolution of design rules for biological automation. Annu. Rev. Biophys. Biomol. Struct. 36, 213–231. doi:10.1146/annurev.biophys.36.040306.132646
Menotti, J., Alanio, A., Sturny-Leclère, A., Vitry, S., Sauvage, F., Barratt, G., et al. (2017). A cell impedance-based real-time in vitro assay to assess the toxicity of amphotericin B formulations. Toxicol. Appl. Pharm. 334, 18–23. doi:10.1016/j.taap.2017.08.017
Mo, S. J., Lee, J., Kye, H. G., Lee, J. M., Kim, E., Geum, D., et al. (2020). A microfluidic gradient device for drug screening with human iPSC-derived motoneurons. Analyst. 145, 3081–3089. doi:10.1039/c9an02384d
Nath, N., Hyun, J., Ma, H., and Chilkoti, A. (2004). Surface engineering strategies for control of protein and cell interactions. Surf. Sci. 570, 98–110. doi:10.1016/j.susc.2004.06.182
Nery, E. W., Jastrzębska, E., Żukowski, K., Wróblewski, W., Chudy, M., and Ciosek, P. (2014). Flow-through sensor array applied to cytotoxicity assessment in cell cultures for drug-testing purposes. Biosens. Bioelectron. 51, 55–61. doi:10.1016/j.bios.2013.07.023
Nguyen, T., Jung, S. H., Lee, M. S., Park, T., Ahn, S., and Kang, J. H. (2019). Robust chemical bonding of PMMA microfluidic devices to porous PETE membranes for reliable cytotoxicity testing of drugs. Lab Chip. 19, 3706–3713. doi:10.1039/c9lc00338j
Ou, X., Chen, P., Huang, X., Li, S., and Liu, B. (2019). Microfluidic chip electrophoresis for biochemical analysis. J. Sep. Sci. 43, 258–270. doi:10.1002/jssc.201900758
Owen, J., Kuznecovs, M., Bhamji, R., William, N., Domenech-Garcia, N., Hesler, M., et al. (2020). High-throughput electrochemical sensing platform for screening nanomaterial–biomembrane interactions. Rev. Sci. Instrum. 91, 025002. doi:10.1063/1.5131562
Pietroiusti, A., Stockmann‐Juvala, H., Lucaroni, F., and Savolainen, K. (2018). Nanomaterial exposure, toxicity, and impact on human health. Wiley Interdiscip. Rev. Nanomed. 10, e1513. doi:10.1002/wnan.1513
Piruska, A., Nikcevic, I., Lee, S. H., Ahn, C., Heineman, W. R., Limbach, P. A., et al. (2005). The autofluorescence of plastic materials and chips measured under laser irradiation. Lab Chip. 5, 1348–1354. doi:10.1039/b508288a
Popova, A. A., Depew, C., Permana, K. M., Trubitsyn, A., Peravali, R., Ordiano, J. A. G., et al. (2017). Evaluation of the droplet-microarray platform for high-throughput screening of suspension cells. SLAS Technol. 22, 163–175. doi:10.1177/2211068216677204
Popova, A. A., Schillo, S. M., Demir, K., Ueda, E., Nesterov‐Mueller, A., and Levkin, P. A. (2015). Droplet‐Array (DA) sandwich chip: a versatile platform for High‐Throughput cell screening based on Superhydrophobic–Superhydrophilic micropatterning. Adv. Mat. 27, 5217–5222. doi:10.1002/adma.201502115
Ramachandran, N., Raphael, J. V., Hainsworth, E., Demirkan, G., Fuentes, M. G., Rolfs, A., et al. (2008). Next-generation high-density self-assembling functional protein arrays. Nat. Methods. 5, 535–538. doi:10.1038/nmeth.1210
Ramirez, C. N., Antczak, C., and Djaballah, H. (2010). Cell viability assessment: toward content-rich platforms. Expert. Opin. Drug. Dis. 5, 223–233. doi:10.1517/17460441003596685
Rashid, A., Vakurov, A., Mohamadi, S., Sanver, D., and Nelson, A. (2017). Substituents modulate biphenyl penetration into lipid membranes. Biochim. Biophys. Acta Biomembr. 1859, 712–721. doi:10.1016/j.bbamem.2017.01.023
Recordati, C., De Maglie, M., Bianchessi, S., Argentiere, S., Cella, C., Mattiello, S., et al. (2015). Tissue distribution and acute toxicity of silver after single intravenous administration in mice: nano-specific and size-dependent effects. Part Fibre Toxicol. 13, 12. doi:10.1186/s12989-016-0124-x
Riahi, R., Shaegh, S. A. M., Ghaderi, M., Zhang, Y. S., Shin, S. R., Aleman, J., et al. (2016). Automated microfluidic platform of bead-based electrochemical immunosensor integrated with bioreactor for continual monitoring of cell secreted biomarkers. Sci. Rep. 6, 1–14. doi:10.1038/srep24598
Ruan, J., Wang, L., Xu, M., Cui, D., Zhou, X., and Liu, D. (2009). Fabrication of a microfluidic chip containing dam, weirs and gradient generator for studying cellular response to chemical modulation. Mat. Sci. Eng. C. 29, 674–679. doi:10.1016/j.msec.2008.12.009
Ruggeri, F. S., Mahul-Mellier, A., Kasas, S., Lashuel, H. A., Longo, G., and Dietler, G. (2017). Amyloid single-cell cytotoxicity assays by nanomotion detection. Cell Death Dis. 3, 1–8. doi:10.1038/cddiscovery.2017.53
Sackmann, E. K., Fulton, A. L., and Beebe, D. J. (2014). The present and future role of microfluidics in biomedical research. Nature. 507, 181–189. doi:10.1038/nature13118
Sarkar, S., Cohen, N., Sabhachandani, P., and Konry, T. (2015). Phenotypic drug profiling in droplet microfluidics for better targeting of drug-resistant tumors. Lab Chip. 15, 4441–4450. doi:10.1039/c5lc00923e
Sarkar, S., Kang, W., Jiang, S., Li, K., Ray, S., Luther, E., et al. (2020). Machine learning-aided quantification of antibody-based cancer immunotherapy by natural killer cells in microfluidic droplets. Lab Chip. 20 (13), 2317–2327. doi:10.1039/D0LC00158A
Shang, W., Liu, Y., Kim, E., Tsao, C., Payne, G. F., and Bentley, W. E. (2018). Selective assembly and functionalization of miniaturized redox capacitor inside microdevices for microbial toxin and mammalian cell cytotoxicity analyses. Lab Chip. 18, 3578–3587. doi:10.1039/c8lc00583d
Shen, S., Zhang, X., Zhang, F., Wang, D., Long, D., and Niu, Y. (2020). Three-gradient constructions in a flow-rate insensitive microfluidic system for drug screening towards personalized treatment. Talanta. 208, 120477. doi:10.1016/j.talanta.2019.120477
Shields, C. W., Reyes, C. D., and López, G. P. (2015). Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip. 15, 1230–1249. doi:10.1039/c4lc01246a
Sochol, R., Sweet, E., Glick, C., Venkatesh, S., Avetisyan, A., Ekman, K., et al. (2016). 3D printed microfluidic circuitry via multijet-based additive manufacturing. Lab Chip. 16, 668–678. doi:10.1039/c5lc01389e
Stoddart, M. J. (2011). Cell viability assays: introduction. Methods. Mol. Biol. 740, 1–6. doi:10.1007/978-1-61779-108-6_1
Strulson, M. K., and Maurer, J. A. (2011). Microcontact printing for creation of patterned lipid bilayers on tetraethylene glycol self-assembled monolayers. Langmuir. 27, 12052–12057. doi:10.1021/la201839w
Sugiura, S., Edahiro, J., Kikuchi, K., Sumaru, K., and Kanamori, T. (2008). Pressure‐driven perfusion culture microchamber array for a parallel drug cytotoxicity assay. Biotechnol. Bioeng. 100, 1156–1165. doi:10.1002/bit.21836
Susloparova, A. A., Koppenhöfer, D., Law, J. K. Y., Vu, X. T., and Ingebrandt, S. (2015). Electrical cell-substrate impedance sensing with field-effect transistors is able to unravel cellular adhesion and detachment processes on a single cell level. Lab Chip. 15, 668–679. doi:10.1039/c4lc00593g
Sweet, E. C., Chen, J. C., Karakurt, I., Long, A. T., and Lin, L. (2017). 3D printed three-flow microfluidic concentration gradient generator for clinical E. coli-antibiotic drug screening. IEEE 30th international conference on micro electro mechanical systems (MEMS), Las Vegas, NV, January 22–26, 2017, (IEEE) 205–208
Ta, V., Park, J., Park, E. J., and Hong, S. (2014). Reusable floating-electrode sensor for the quantitative electrophysiological monitoring of a nonadherent cell. ACS Nano. 8, 2206–2213. doi:10.1021/nn4053155
Tran, T. B., Cho, S., and Min, J. (2013). Hydrogel-based diffusion chip with electric cell-substrate impedance sensing (ECIS) integration for cell viability assay and drug toxicity screening. Biosens. Bioelectron. 50, 453–459. doi:10.1016/j.bios.2013.07.019
Urcan, E., Haertel, U., Styllou, M., Hickel, R., Scherthan, H., and Reichl, F. X. (2010). Real-time xCELLigence impedance analysis of the cytotoxicity of dental composite components on human gingival fibroblasts. Dent. Mater. J. 26, 51–58. doi:10.1016/j.dental.2009.08.007
Vaidyanathan, R., Yeo, T., and Lim, C. T. (2018). Microfluidics for cell sorting and single cell analysis from whole blood. Methods Cell Biol. 147, 151–173. doi:10.1016/bs.mcb.2018.06.011
Vajrala, V. S., Belaïdi, F. S., Lemercier, G., Zigah, D., Rigoulet, M., Devin, A., et al. (2019). Microwell array integrating nanoelectrodes for coupled opto-electrochemical monitorings of single mitochondria. Biosens. Bioelectron. 126, 672–678. doi:10.1016/j.bios.2018.11.036
Wang, G., Dewilde, A. H., Zhang, J., Pal, A., Vashist, M., Bello, D., and Therrien, J. M. (2011). A living cell quartz crystal microbalance biosensor for continuous monitoring of cytotoxic responses of macrophages to single-walled carbon nanotubes. Part Fibre Toxicol. 8, 4. doi:10.1186/1743-8977-8-4
Wang, T., Liu, J., and Nie, F. (2018). Non-dye cell viability monitoring by using pH-responsive inverse opal hydrogels. J. Mater. Chem. B. 6, 1055–1065. doi:10.1039/c7tb02631e
Wang, Z., Kim, M., Marquez, M., and Thorsen, T. (2007). High-density microfluidic arrays for cell cytotoxicity analysis. Lab Chip. 7, 740–745. doi:10.1039/b618734j
Wlodkowic, D., Faley, S., Zagnoni, M., Wikswo, J. P., and Cooper, J. M. (2009). Microfluidic single-cell array cytometry for the analysis of tumor apoptosis. Anal. Chem. 81, 5517–5523. doi:10.1021/ac9008463
Wu, W., Shen, J., Banerjee, P., and Zhou, S. (2010). Core–shell hybrid nanogels for integration of optical temperature-sensing, targeted tumor cell imaging, and combined chemo-photothermal treatment. Biomaterials. 31, 7555–7566. doi:10.1016/j.biomaterials.2010.06.030
Wurm, M., Schöpke, B., Lutz, D., Müller, J., and Zeng, A. (2010). Microtechnology meets systems biology: the small molecules of metabolome as next big targets. J. Biotechnol. 149, 33–51. doi:10.1016/j.jbiotec.2010.05.002
Xia, Y., and Whitesides, G. M. (1998). Soft lithography. Annu. Rev. Mater. Sci. 28, 153–184. doi:10.1146/annurev.matsci.28.1.153
Xing, J. Z., Zhu, L., Jackson, J. A., Gabos, S., Sun, X., Wang, X., et al. (2005). Dynamic monitoring of cytotoxicity on microelectronic sensors. Chem. Res. Toxicol. 18, 154–161. doi:10.1021/tx049721s
Yu, L., Chen, M. C., and Cheung, K. C. (2010). Droplet-based microfluidic system for multicellular tumor spheroid formation and anticancer drug testing. Lab Chip. 10, 2424–2432. doi:10.1039/c004590j
Zhang, D., Peng, Y., Qi, H., Gao, Q., and Zhang, C. (2010). Label-free electrochemical DNA biosensor array for simultaneous detection of the HIV-1 and HIV-2 oligonucleotides incorporating different hairpin-DNA probes and redox indicator. Biosens. Bioelectron. 25, 1088–1094. doi:10.1016/j.bios.2009.09.032
Zhang, H., Lee, Y. Y., Leck, K. J., Kim, N. Y., and Ying, J. Y. (2007). Recyclable hydrophilic− hydrophobic micropatterns on glass for microarray applications. Langmuir. 23, 4728–4731. doi:10.1021/la063759i
Zhang, L., Chen, P., Zhou, Z., Hu, Y., Sha, Q., Zhang, H., et al. (2019). Agarose-based microwell array chip for high-throughput screening of functional microorganisms. Talanta. 191, 342–349. doi:10.1016/j.talanta.2018.08.090
Zhang, S., Nelson, A., and Beales, P. A. (2012). Freezing or wrapping: the role of particle size in the mechanism of nanoparticle–biomembrane interaction. Langmuir. 28, 12831–12837. doi:10.1021/la301771b
Zhao, L., Cao, J., Wu, Z., Li, J., and Zhu, J. (2013). Lab-on-a-chip for anticancer drug screening using quantum dots probe based apoptosis assay. J. Biomed. Nanotechnol. 9, 348–356. doi:10.1166/jbn.2013.1546
Zhao, L., Cheng, P., Li, J., Zhang, Y., Gu, M., Liu, J., et al. (2009). Analysis of nonadherent apoptotic cells by a quantum dots probe in a microfluidic device for drug screening. Anal. Chem. 81, 7075–7080. doi:10.1021/ac901121f
Zhao, Y., Cheng, Y., Shang, L., Wang, J., Xie, Z., and Gu, Z. (2015). Microfluidic synthesis of barcode particles for multiplex assays. Small. 11 (2), 151–174. doi:10.1002/smll.201401600
Keywords: microtechnology, lab-on-a-chip, cytotoxicity, surface engineering, microfluidics, Sensors, Biosensors, single cell
Citation: Garcia-Hernando M, Benito-Lopez F and Basabe-Desmonts L (2020) Advances in Microtechnology for Improved Cytotoxicity Assessment. Front. Mater. 7:582030. doi: 10.3389/fmats.2020.582030
Received: 14 July 2020; Accepted: 26 October 2020;
Published: 20 November 2020.
Edited by:
Patricia Krawczak, IMT Lille Douai, FranceReviewed by:
M. Gabriella Santonicola, Sapienza University of Rome, ItalyHuaiyu Wang, Chinese Academy of Sciences (CAS), China
Copyright © 2020 Garcia-Hernando, Benito-Lopez and Basabe-Desmonts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lourdes Basabe-Desmonts, lourdes.basabe@ehu.eus Fernando Benito-Lopez, fernando.benito@ehu.eus
 Maite Garcia-Hernando1,2
Maite Garcia-Hernando1,2  Lourdes Basabe-Desmonts
Lourdes Basabe-Desmonts