Wooden Stepping Stones: Diversity and Biogeography of Deep-Sea Wood Boring Xylophagaidae (Mollusca: Bivalvia) in the North-East Atlantic Ocean, With the Description of a New Genus
- 1Centre d’Estudis Avançats de Blanes (CEAB-CSIC), Blanes, Spain
- 2Max Planck Institute for Marine Microbiology, Bremen, Germany
- 3Sorbonne Université, CNRS, Observatoire Océanologique de Banyuls (LECOB), Banyuls-sur-Mer, France
- 4Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA, United States
Wood boring bivalves of the family Xylophagaidae inhabit sunken wood on the deep-sea floor where they play a key role in the degradation of this organic matter in the ocean. The patchiness of wood-fall habitats is impeding targeted sampling and little is therefore known on xylophagaid biology. We investigated for the first time the diversity and biogeography of Xylophagaidae in the NE-Atlantic and the Mediterranean over a broad geographic range and in various water depths using experimental wood deployments. We combined morphological and molecular analyses for species discrimination. A phylogenetic reconstruction based on 18S and 28S rRNA and COI genes revealed non-monophyly of the type genus, Xylophaga Turton (1822), and led us to revise the taxonomy and erect the genus Xylonora gen. nov. COI haplotypes of the most abundant species revealed broad Atlanto-Mediterranean genetic connectivity for Xylophaga dorsalis and Xylonora atlantica new comb., while genetic connectivity appears limited for Abditoconus brava across the entrance of the Mediterranean. We provide the first COI barcode data for Xylophagaidae as a solid base for future taxonomic work. Wood deployments in a broad geographic range provided a powerful tool for research on Xylophagaidae allowing for conclusions on ecological requirements of xylophagaid species.
Introduction
The Xylophagaidae Purchon (1941) and the sister family Teredinidae Rafinesque (1815) are among the most abundant wood-degrading organisms in the marine realm (Turner, 1973; Wolff, 1979; Distel, 2003; Distel et al., 2011). Both bivalve families have similar habitats, as they both bore in wood which they use as a shelter and they are both considered obligate wood feeders with limited ability for filter feeding (Purchon, 1941; Saraswathy and Nair, 1971). Nevertheless, their ecological niches do not overlap. Xylophagaidae live in deep waters where they colonize wood that has sunk to the sea floor. Teredinidae, known also as shipworms, colonize driftwood in surface waters. Shipworms gained a reputation as pests by damaging boats and submerged wooden structures ever since humans started to sail, and their life histories and growth rates are therefore well studied (Haderlie and Mellor, 1973; Culliney, 1975; MacIntosh et al., 2014). Xylophagaidae in contrast, have been recorded mostly below 150 m depth down to hadal environments at 7,000 m depth (Knudsen, 1961; Turner, 2002; Distel, 2003; Voight, 2008) and many aspects of their life histories remain unknown.
Xylophagaids and teredinids ingest wood chips they create by boring with their specialized shells and which then accumulate in a special stomach diverticulum, the caecum (Purchon, 1941; Monari, 2009; Distel et al., 2011). Cellulose, hemicellulose and lignin, the main wood components, are highly refractory polysaccharide polymers that do not hydrolyse easily. Biological degradation uses cellulolytic enzymes that break them down to easily digestible mono- and disaccharides (Klemm et al., 2005). Many bacteria and fungi synthesize cellulolytic enzymes, while most animals do not. Teredinids harbor cellulose-degrading bacteria in their gills (Waterbury et al., 1983; Distel et al., 2002; Distel, 2003) and this symbiosis is considered as the basis for their wood digestion capacity (O’Connor et al., 2014). However, an additional combination of cellulolytic enzymes produced by the molluscs themselves has been recently discovered in the caecum (Sabbadin et al., 2018). Xylophagaidae also host endosymbiotic bacteria in the gills (Distel and Roberts, 1997; Bessette et al., 2014; Fagervold et al., 2014; Voight, 2015), but their role and metabolic pathways remain even less studied.
Wood reaches the open ocean by river transport after storm and flood events (Stockton and DeLaca, 1982; West et al., 2011), and after drifting and becoming saturated with water it eventually sinks down to the seafloor (Wolff, 1979). Sunken wood represents high local carbon inputs to the deep sea which generally lacks primary production and receives extremely limited phototrophic contributions from the surface (Stockton and DeLaca, 1982; Smith et al., 2008). Wood-falls create discrete organic islands on the deep-sea floor harboring distinct and specialized fauna mining the wood, breaking it up and thereby supporting wood polymer degradation by cellulolytic microorganisms (Kalenitchenko et al., 2018b). Consumption of oxygen during this process eventually leads to anoxic and sulfidic environments providing habitats for chemosynthetic microbial communities and invertebrates associated with sulfur-oxidizing symbionts (Gaudron et al., 2010; Bienhold et al., 2013; Fagervold et al., 2013; Yücel et al., 2013; Kalenitchenko et al., 2018a).
Xylophagaids are primarily responsible for the structural degradation of wood in the deep sea (Turner, 1977) building up very dense populations within a few weeks to several months (Romey et al., 1994; Harvey, 1996; Tyler et al., 2007; Romano et al., 2013). Their fast-growing populations become a valuable trophic resource for carnivores, while their fecal materials play a similar role for detritivores. This is certainly a basal shackle of the trophic chain in deep-sea wood-fall ecosystems which makes xylophagaids keystone for the associated assemblages (Turner, 1973; Culliney and Turner, 1976; Distel, 2003).
Deep-sea wood falls are highly fragmented and ephemeral sulfidic habitats comparable to other chemosynthetic ecosystems (e.g., whale falls, hydrothermal vents, hydrocarbon seeps), but they sustain even more rapid ecological cycles (Kalenitchenko et al., 2018b). Organisms living in such patchy habitats often develop long-distance dispersal strategies to colonize areas hundreds of kilometers away (Baco et al., 1999; Tyler and Young, 1999; Van Dover et al., 2002; Breusing et al., 2016). Studies on xylophagaids relied for long on occasionally recovered naturally sunken wood, mainly as by-catch in trawl and dredge hauls. Collections of xylophagaids remained largely untargeted and very little is known on their distribution mechanisms and geographical patterns. A few species have been collected repeatedly at different sites, such as Xylophaga dorsalis (Turton, 1819) and Xylophaga atlantica Richards, 1942. Both live in wide geographic areas and from littoral to bathyal depths in the northern Atlantic Ocean (Purchon, 1941; Culliney and Turner, 1976; Santhakumaran, 1980; Voight, 2007). Many other species are only known from single records (Knudsen, 1961; Harvey, 1996; Turner, 2002; Voight, 2007, 2008), while some occur in abyssal plains or even in deep-sea trenches, thousands of kilometers away from the nearest land mass, where sunken wood is considered extremely rare (Knudsen, 1961; Voight, 2008; Voight and Segonzac, 2012).
Xylophagaidae currently includes six genera: Xylophaga Turton, 1822, Xylopholas Turner, 1972, Xyloredo Turner, 1972, Feaya Voight, 2019, Abditoconus Voight, 2019, and Spiniapex Voight, 2019 (Voight et al., 2019). Xylophaga is the most speciose, containing currently more than 50 species. Historically, the vast majority of taxonomic works on Xylophagaidae have based on morphology, while only two studies also used molecular markers; hence little is currently known about their genetic diversity (Romano et al., 2014; Voight et al., 2019). The main diagnostic morphological characters refer to valves, mesoplax, siphons, and muscle inserts on inner surfaces of the valves (Tyler et al., 2007). These characters and the overall shape of the shells may vary among populations and are affected by environmental conditions such as wood hardness. Even individuals within the same population inhabiting neighboring logs of different wood such as soft pine or hard oak may vary (Turner, 2002). In addition, extraction of the xylophagaids from wood substrates can be challenging and often lead to fragmentation of specimens and damage of their delicate shells. As a result, several species descriptions are based on single or incomplete specimens, lacking information on intraspecific phenotypic variation (Smith, 1903; Knudsen, 1961; Santhakumaran, 1980; Harvey, 1996; Turner, 2002). For these reasons, species determinations relying only on morphological characters can be ambiguous and lead to errors. Molecular tools can circumvent many of these difficulties, and they are particularly useful for increasing rigor of identification, especially in poorly known species.
Recent in situ deep-sea experiments used moorings, submersibles or remotely operated vehicles for deploying wood at great depths. These experiments were designed to specifically target wood-fall organisms, including xylophagaids. Previous studies mainly focused on the eastern Pacific and western Atlantic and very little is currently known on the biogeographic distribution of the European species (Tyler et al., 2007; Voight, 2007, 2008; Reft and Voight, 2009; Voight and Segonzac, 2012). However, several European oceanographic research projects recently deployed wood in the North-East Atlantic and Mediterranean deep sea allowing for comprehensive studies on European Xylophagaidae (Gaudron et al., 2010, 2016; Bienhold et al., 2013; Romano et al., 2013, 2014).
In this study we investigated the diversity and biogeographic distribution of Xylophagaidae in a broad range of boreal to temperate latitudes in North-East Atlantic and Mediterranean waters based on numerous experimental wood deployments. We used molecular methods and analyzed for the first time cytochrome c oxidase subunit I (COI) data of Xylophagaidae from a depth range of 130 to 2,300 m to verify morphology-based species identification. The aims of the present study were to (1) assess the diversity and phylogeny of these particular organisms, (2) provide barcode data, (3) reveal genetic connectivity patterns for populations of Xylophagaidae, and (4) draw up conclusions on ecological requirements of xylophagaid species.
Materials and Methods
Sampling and Specimens Collection
Wood samples were experimentally deployed on the deep-sea floor and recovered several months later during a number of oceanic research cruises between 2006 and 2013 in various sites in the North-East Atlantic Ocean and the Mediterranean.
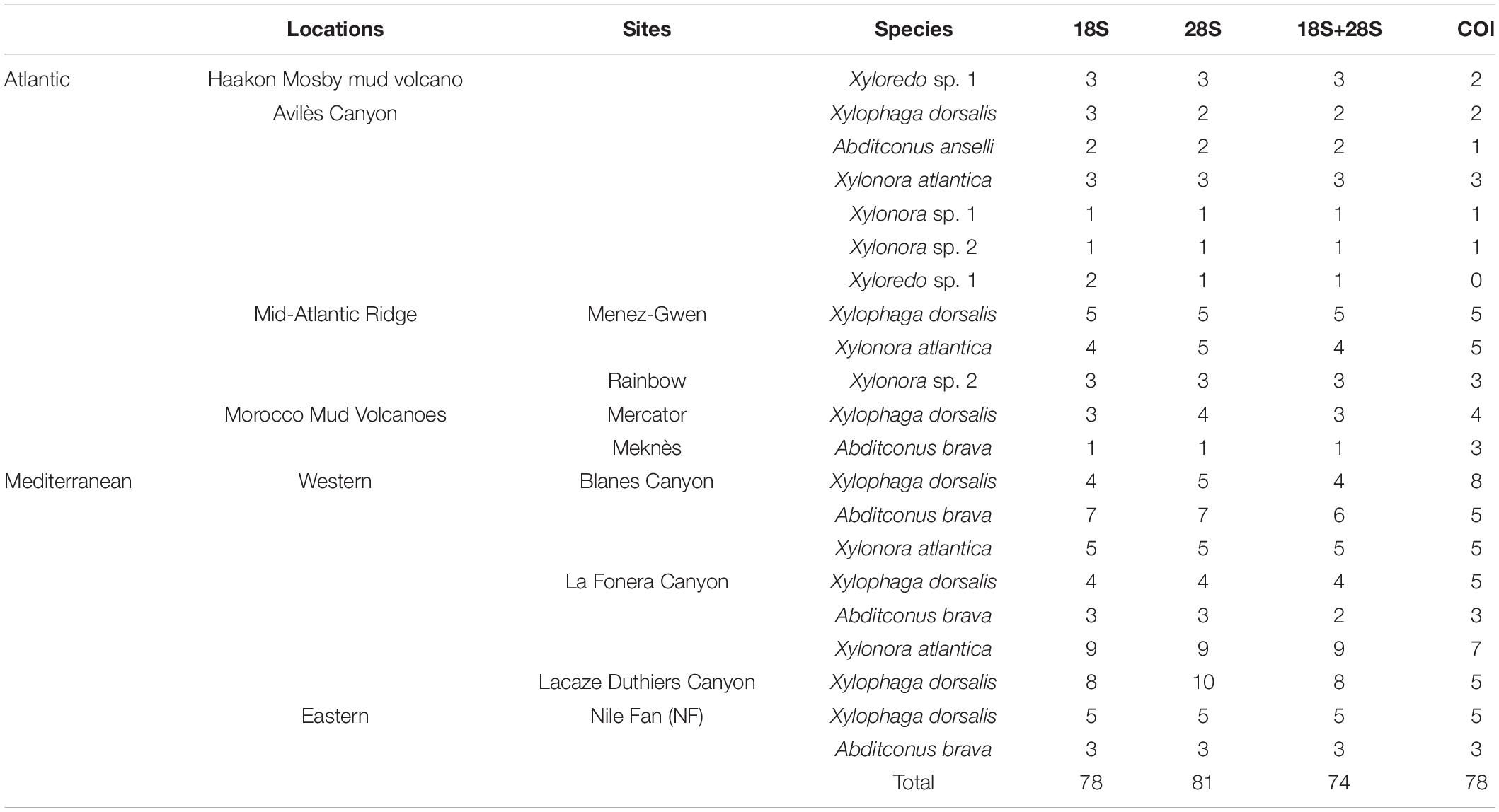
Experimental wood deployments in the Atlantic included the Haakon Mosby Mud Volcano (HMMV) in the deep Barents Sea, the Avilés submarine canyon (AC) in the Bay of Biscay, the hydrothermal vent sites Rainbow (Rb) and Menez Gwen (MG) on the Mid-Atlantic Ridge (MAR) and the Meknès and Mercator mud volcanoes in the North Atlantic off Morocco (Morocco mud volcanoes MMV). The Mediterranean sites included three submarine canyons offshore north-eastern Iberian Peninsula and southern France: Blanes (BC), La Fonera (LFC) and Lacaze-Duthiers (LDC) (North Western Mediterranean), and one cold seep site (Central Pockmarks) in the deep Nile Fan (NF) off Egypt (Eastern Mediterranean) (Figure 1 and Table 1). Deployments ranged from 130 to 2,300 m depth. Wood parcels were either placed directly on the deep-sea floor (Figure 2) or moored 20 to 30 m above. At hydrothermal vent fields and mud volcanoes, wood was placed away from the influence of active hot fluid emissions or seepage. The experiments used soft (pine and Douglas fir) or hard (oak) wood, either as cubes within colonization devices (Gaudron et al., 2010; Cunha et al., 2013), free logs (Bienhold et al., 2008, 2013) or logs inside net bags (Romano et al., 2013, 2014; Kalenitchenko et al., 2018b). Wood deployment times ranged from 3 to 36 months.
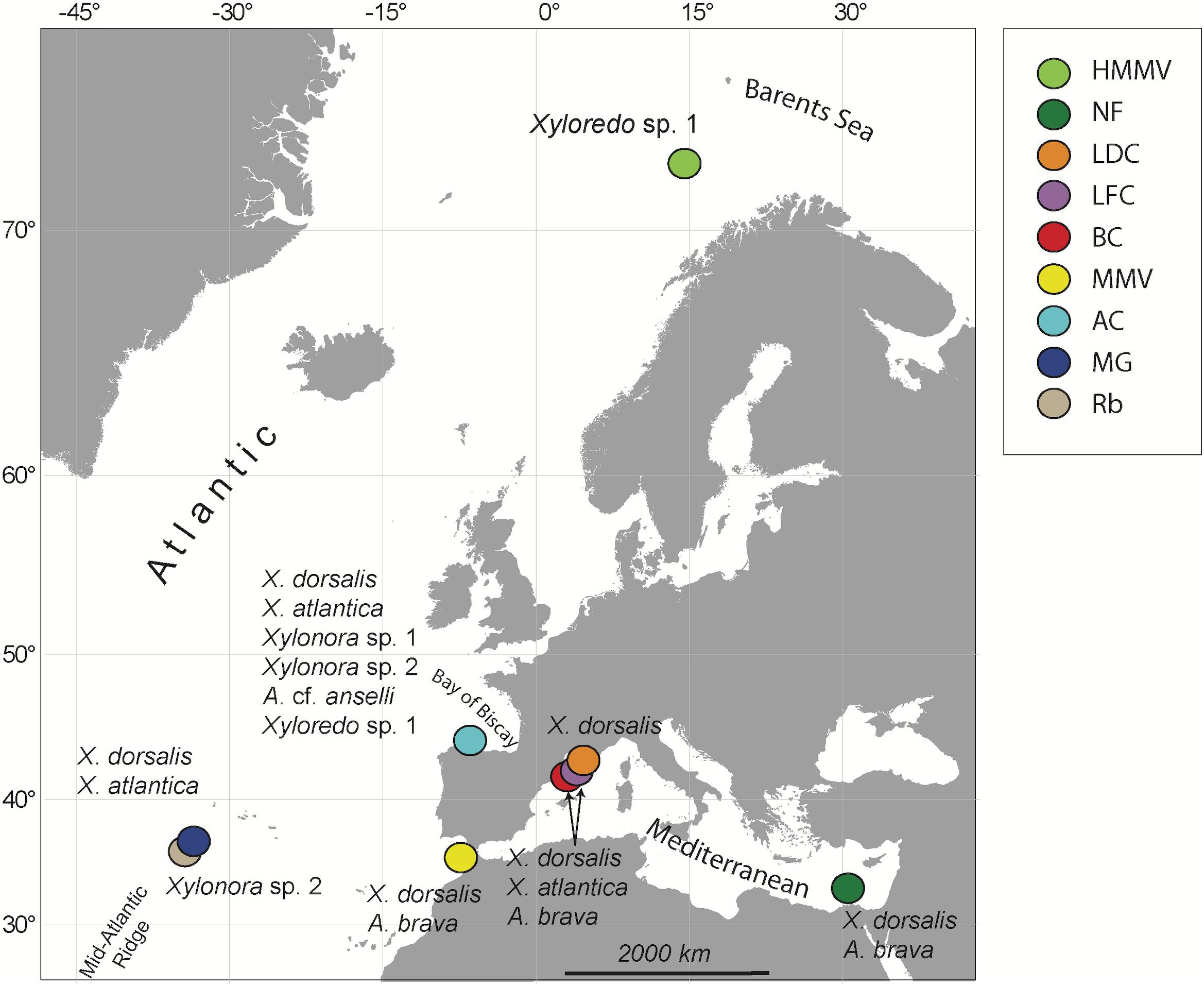
Figure 1. Sites of experimental wood deployments. HMMV, Haakon Mosby mud volcano; NF, Nile Fan; LDC, Lacaze-Duthiers Canyon; LFC, La Fonera Canyon; BC, Blanes Canyon; MMV, Morocco mud volcanoes, including Mercator and Meknès mud volcanoes; AC, Avilés Canyon; RB, Rainbow; and MG, Menez Gwen hydrothermal vent fields on the Mid-Atlantic Ridge. Recovered species of Xylophagaidae are shown close to each site.
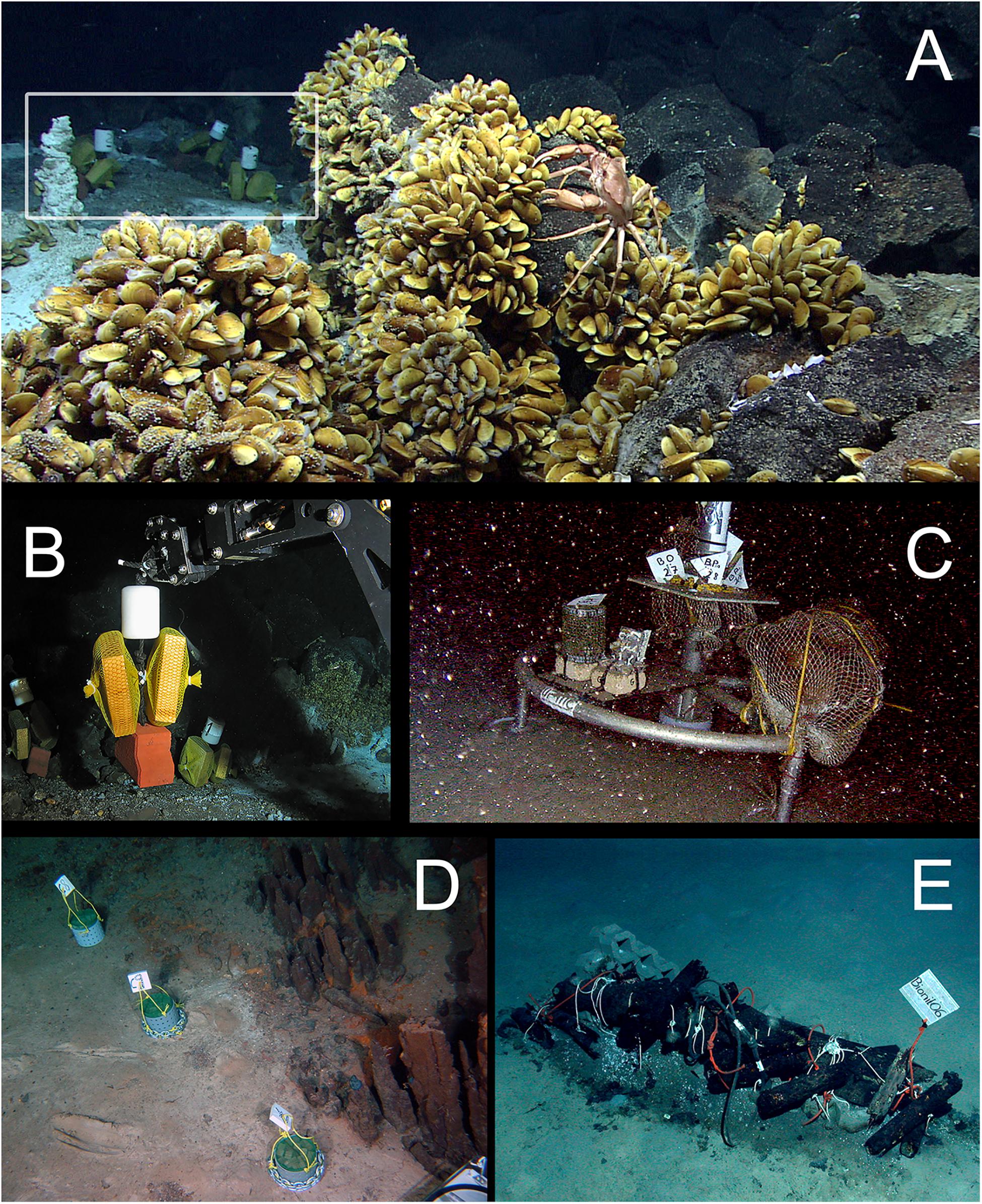
Figure 2. Examples of in situ wood deployments. (A) Small wood panels (white frame) deposited outside of a diffuse flow area in the Menez Gwen hydrothermal vent field (RV Meteor, cruise M82/3); (B) ROV manipulator arm positioning wood panels in panel (A) at Menez Gwen (RV Poseidon, cruise POS402); (C) Moored lander with net bags containing wood blocks in Lacaze-Duthiers Canyon (LECOB LDC monitoring cruises); (D) Chemecoli colonization devices containing small pine wood cubes (2.5 cmł) next to inactive chimneys in the Rainbow hydrothermal vent field (RV Pourquoi Pas?, cruise Momardream); (E) Chemecoli colonization devices and large wood logs in the deep Nile Fan (RV Pourquoi Pas?, cruise Medeco). Copy rights: (A,B,E) MARUM–Center for Marine Environmental Sciences, University of Bremen; (C) Sorbonne Université; (D) Ifremer.
The recovered wood pieces were opened using hammer and chisel or, when degradation had considerably progressed, by hand. The bivalves were carefully extracted and preserved for morphological identification in a 4% paraformaldehyde seawater solution, then transferred to ethanol/sterile filtered seawater solution at v/v 70%. For molecular analyses, they were either directly frozen at −20°C or preserved in 99% ethanol. Before molecular analyses, the specimens were sorted under a dissecting microscope and identified to species or morphotype level, based on the relevant literature (Harvey, 1996; Turner, 2002; Voight, 2007, 2008; Romano et al., 2014).
DNA Extraction, PCR Amplification and Sequencing
We extracted DNA from dissected fragments of siphon or foot tissues from 92 specimens (Table 2), following either a modified protocol based on (Zhou et al., 1996) or with a DNeasy Mini Kit (Qiagen)1. Nuclear 18S and 28S rRNA genes were amplified by polymerase chain reaction (PCR) using seven primer pairs (Distel et al., 2011; Supplementary Table S1) and by using various enzymes and amplification conditions (Supplementary Table S2). A fragment of mitochondrial COI was amplified with a combination of the general primer HCO2198 (Folmer et al., 1994) and an unpublished primer developed by Dario Zuccon (National Natural History Museum of Paris, pers.com.), generating a size of ∼300–500 bp (Supplementary Table S1). Amplification reactions were conducted with 1 to 2 μl of DNA extract in final volumes of 20 or 50 μl, following the enzyme provider recommendations. PCR products were purified with the QIAquick kit (Qiagen, Hilden, Germany) or ExoSAP product Clean-up.
Bidirectional direct sequencing using the same primers as for PCR (Supplementary Table S1) and additional internal primers (Supplementary Table S1) was done with the Bigdye v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, United States) and the sequencer ABI prism 3130×L (Applied Biosystems, Foster City, UA, United States) or by Macrogen (Korea).
Sequence Analyses and Phylogenetic Reconstructions
The sequences were edited in Geneious 8.1.8 (Kearse et al., 2012) and overlapping fragments merged into consensus sequences. We compared our sequences with the NCBI databases using BLAST (Altschul et al., 1997) to check for sequence contaminations (Vrijenhoek et al., 2009). The NCBI database did not contain COI sequences from Xylophagaidae and the similarity hits targeted COI sequences of unrelated bivalve and gastropod molluscs (max. 70% sequence similarity). COI sequences were translated into amino acid alignment with code 5 for invertebrate mitochondrial genes and checked for stop codons in order to avoid pseudogenes. NCBI comparisons of large and small rRNA subunit sequences revealed highest similarities with species of Xylophagaidae and Teredinidae (max. 98% for 18S rRNA, 84% for 28S rRNA).
We imported our sequences in Mesquite 3.2 (Maddison and Maddison, 2019) and included NCBI sequences of Xylophagaidae, Teredinidae, and Pholadidae (Supplementary Table S3). All sequences were aligned using MAFFT and then manually edited, trimmed and cleaned from ambiguous positions. The final alignments included 388 bp (COI), 1353 bp (18S rRNA), and 1104 bp (28S rRNA). All new sequences are deposited on GenBank (Supplementary Table S3).
Genetic diversity was evaluated as the number of segregating sites (S), using the MEGA software version 6 (Tamura et al., 2013) and uncorrected pairwise distances with PAUP∗ v.4.0a161 (Swofford, 2003).
Maximum likelihood (ML) and Bayesian inference (BI) analyses of the molecular data phylogenetic trees were calculated separately for each gene COI, 18S rRNA and 28S rRNA, using RAxML v.8.2.10 (Stamatakis, 2014) and MrBayes v.3.2.6 (Huelsenbeck and Ronquist, 2001), respectively. Analyses of concatenated COI+18S rRNA+28S rRNA, partitioned by gene, were also calculated with RAxML and MrBayes. For the BI analyses, the best-fitting evolutionary model for each gene was estimated with the software package Iq-Tree 1.6.12 (Chernomor et al., 2016) using the Akaike information criterion (AIC). GTR+F+I+G4 was the best fitting nucleotide substitution model for each of the genes. BI analyses were run twice, starting from random trees, with four chains running simultaneously (two cold and two heated). Chains were run for 106 generations for 18S rRNA and 28S rRNA and 107 generations for COI and the concatenated data, sampled every 1,000 generations and, after verifying that stationarity had been reached, 25% of the generations were discarded as burn-in. The ML analyses were performed using the GTR+G evolutionary model, following the recommendations in the software author. Support values for the ML analyses were generated by 100 bootstrap pseudoreplicates. We used sequences from Mya arenaria Linnaeus, 1758, Mya truncata Linnaeus, 1758 and Varicorbula limatula Conrad, 1846 [here reported as Varicorbula disparilis D’Orbigny, 1853 as in NCBI database] and the Teredininae Lyrodus pedicellatus Quatrefages, 1849 and Bankia carinata Gray, 1827 as outgroups. Trees were visualized in FigTree v 1.4.2 (Rambaut, 2006). A median-joining haplotype network based on COI sequences was constructed in PopArt (Leigh and Bryant, 2015) for the species that produced a minimum of 15 sequences, i.e., X. dorsalis, Abditoconus brava, and X. atlantica.
Nomenclatural Acts
This article conforms to the requirements of the amended International Code of Zoological Nomenclature (ICZN), and hence the new names contained herein are available under that Code from the electronic edition of this article. This published work including the nomenclatural acts have been registered in the ICZN online registration system ZooBank. The ZooBank Life Science Identifiers (LSID) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/. The LSID for this publication is: urn:lsid:zoobank.org:pub:71C8DF63-5C84-4C6E-B5A2-7902939FEBBD.
Results
Phylogenetic Analyses
The phylogenetic trees obtained with both tree reconstruction methods for the single genes partially resolved the phylogeny. The best resolution was achieved in the combined analysis of all three genes and the BI and ML analyses provided consistent topologies in this respect. We show the BI tree for the three concatenated genes, including the BI posterior probabilities (pp) and the ML bootstrap (bs) on the nodes (Figure 3).
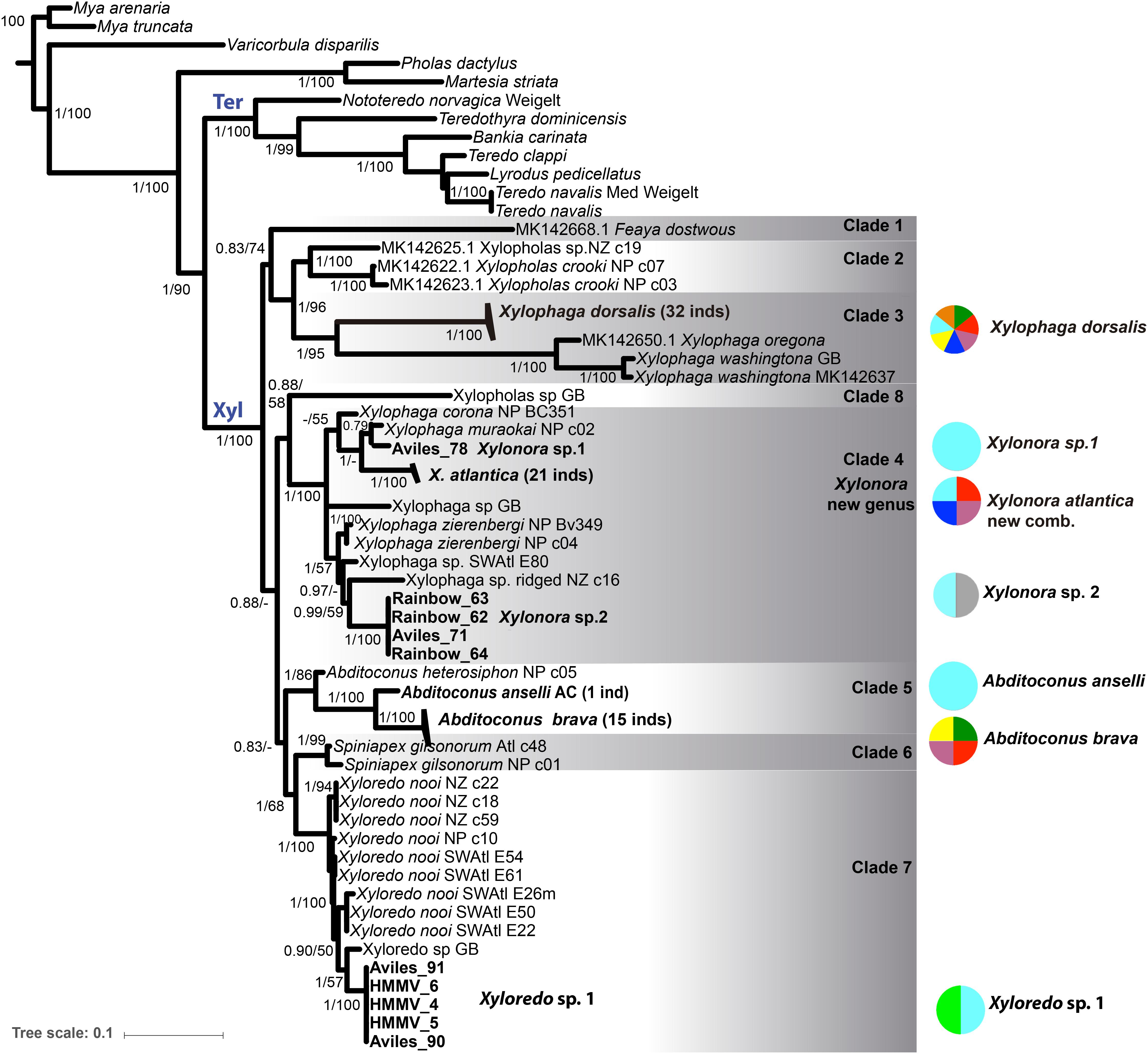
Figure 3. Bayesian inference (BI) and Maximum likelihood (ML) phylogenetic analysis based on the concatenated data set of COI, 18S and 28S rRNA genes (MAFFT aligned). Statistic support is indicated on the nodes (BI/ML, values < 0.5 pp or 50% bs not shown). Bold taxa names: sequences from this study, including locations. Not-bold taxa names: GenBank sequences; inds = number of sequenced individuals. (see Supplementary Table S3 for accession numbers). Xyl: node of Xylophagaidae; Ter: node of Teredinidae. Colored circles and associated numbers represent the geographic and depth distributions of the species in this study; circle colors as in Figure 1.
Xylophagaidae formed a well-supported clade (1 pp, 100% bs), more closely related to Teredinidae than to Pholadidae (Figure 3). Eight “major” clades were recovered within the family, six of them were well supported (Figure 3, >0.9 pp, >70% bs) and two of them (clades 1 and 8) had weak support, similar to the result in Voight et al. (2019). Clade 1 was formed by a single lineage originally included in Xylopholas (Voight, 2016), but recently moved to Feaya (Voight et al., 2019), and this species was sister group to clades 2 and 3, with moderate support. Xylophaga was non-monophyletic, with its species distributed over clades 3 and 4. Clade 3 comprised the type species, X. dorsalis, Xylophaga oregona Voight (2007) and also Xylophaga washingtona Bartsch (1921), all three morphologically characterized by truncated excurrent siphons [groups 5 and 6 in (Turner, 2002)]. Their sister-clade relationship to clade 2, which combines recently sequenced specimens of Xylopholas (Voight et al., 2019), was well supported. Clade 4 contained other Xylophaga species and was well-supported. To resolve the non-monophyly of Xylophaga, we propose to assign clade 4 to a new genus, namely Xylonora gen. nov., since the type species of Xylophaga, X. dorsalis, is member of clade 3. The species of Abditoconus were united in clade 5, while those of Spiniapex were in clade 6, and those of Xyloredo in clade 7. A single terminal, supposedly a Xylopholas from the Gulf of Mexico, represented by GenBank sequences (Clade 8, 0.9 pp, 58% bp), was isolated from all other Xylopholas. This situation is unclear and could possibly result from misidentification of the specimen. Re-collection and examination of the voucher may be needed to solve this in the future.
In summary, the monophyletic and well-supported xylophagaid clades correspond to six genera Xylonora gen. nov., Xylopholas, Xylophaga, Abditoconus, Spiniapex, and Xyloredo, as well as the monotypic Feaya.
Taxonomic Account
Xylophaga Turton, 1822 sensu stricto
Type species
Xylophaga dorsalis Turton, 1819.
Diagnosis
Mesoplax with two calcareous plates, excurrent siphon truncated, shorter than incurrent siphon, periostracum restricted to basal and lateral siphon, and a furrow on dorsal incurrent siphon distal to excurrent opening.
Species included
Three groups of species are included in the genus at present. The first includes X. dorsalis, X. washingtona, and X. oregona (molecular data of these three species included in this study, Supplementary Table S3). For the second group, molecular data are not yet available: Xylophaga alexisi Voight and Segonzac, 2012, Xylophaga aurita Knudsen, 1961, Xylophaga bayeri Turner, 2002, Xylophaga depalmai Turner, 2002; Xylophaga guineensis Knudsen, 1961, Xylophaga indica Smith, 1904, Xylophaga mexicana Dall, 1908, Xylophaga multichela Voight, 2008, Xylophaga nidarosiensis Santhakumaran, 1980, Xylophaga praestans Smith, 1903, Xylophaga pacifica Voight, 2009, Xylophaga rikuzenica Taki and Habe, 1945, Xylophaga turnerae Knudsen, 1961, Xylophaga tipperi Turner, 2002. For the third group, additional morphologic or genetic information are needed: Xylophaga africana Knudsen, 1961, Xylophaga lobata Knudsen, 1961, Xylophaga obtusata Knudsen, 1961, Xylophaga clenchi Turner and Culliney, 1971, Xylophaga noradi Santhakumaran, 1980, Xylophaga tubulata Knudsen, 1961 Xylophaga whoi Turner, 2002, Xylophaga grevei, Xylophaga ricei Harvey, 1996, and Xylophaga gagei Harvey, 1996.
Remarks
The morphology of the species of the third group fits better to the diagnosis of the new genus Xylonora as their excurrent siphons are equal to subequal with respect to the incurrent siphons rather than truncated as in Xylophaga. However, additional morphologic or genetic information is needed before suggesting a transfer to this genus.
Xylonora gen. nov. Romano
LSID: urn:lsid:zoobank.org:act:9D8F449F-8720-4E9C-9814-FCA1292AC0F2.
Type species
Xylonora atlantica (Richards, 1942) new comb.
Diagnosis
Mesoplax with two calcareous plates of variable shape; siphons equal or subequal; excurrent siphon opening dorsally to incurrent one; cirri on both openings present or absent. Small individuals (embryos or dwarf males) may occur attached dorsally on shell, posterior to umbo but seldom to ventral siphon.
Species included
Xylonora atlantica new comb., Xylonora muraokai Turner, 2002 new comb., Xylonora corona Voight, 2007 new comb. and Xylonora zierenbergi Voight, 2007 new comb. (molecular data included in this study, Supplementary Table S3).
Etymology
The genus is named in honor of CR’s mother Eleonora “Nora” Rivarola, Professor of Chemistry, who always provided inspiration, rigor and motivation as role model for women in science.
Remarks
Our molecular results are consistent with the morphology, showing clade 4 species clearly separated from Xylophaga sensu stricto (i.e., clade 3), which have excurrent siphons much shorter than incurrent ones (instead of equal or subequal in Xylonora gen. nov.), lateral lobes along the dorsal surface of incurrent siphons (absent in Xylonora gen. nov.) and large fecal chimney around the siphon (absent in Xylonora gen. nov.). This new genus may include up to 33 nominal species if additional species were to be transferred from Xylophaga. This genus would then correspond to species groups 1 to 4 in Turner (2002).
Similar to Xylopholas scrippsorum Voight, 2009, in some species of Xylonora gen. nov. small individuals or dwarf males attach to the ventral mantle or laterally or ventrally to the adult siphons. The position of the small individuals on the adults is not a distinguishing character for Xylonora gen. nov. They have been found dorsally attached on adults in Xylonora atlantica new comb., Xylophaga africana, Xylophaga clenchi, Xylophaga lobata, Xylophaga obtusata, Xylophaga panamensis Knudsen, 1961, but sometimes attached to ventral siphon like in Xylophaga concava Knudsen, 1961, Xylophaga tubulata Knudsen, 1961, Xylophaga wolffi-grevei Knudsen, 1961, and X. dorsalis. However the presence of small individuals attached to adults may have been overlooked in other species or genera.
Species Diversity in European Waters
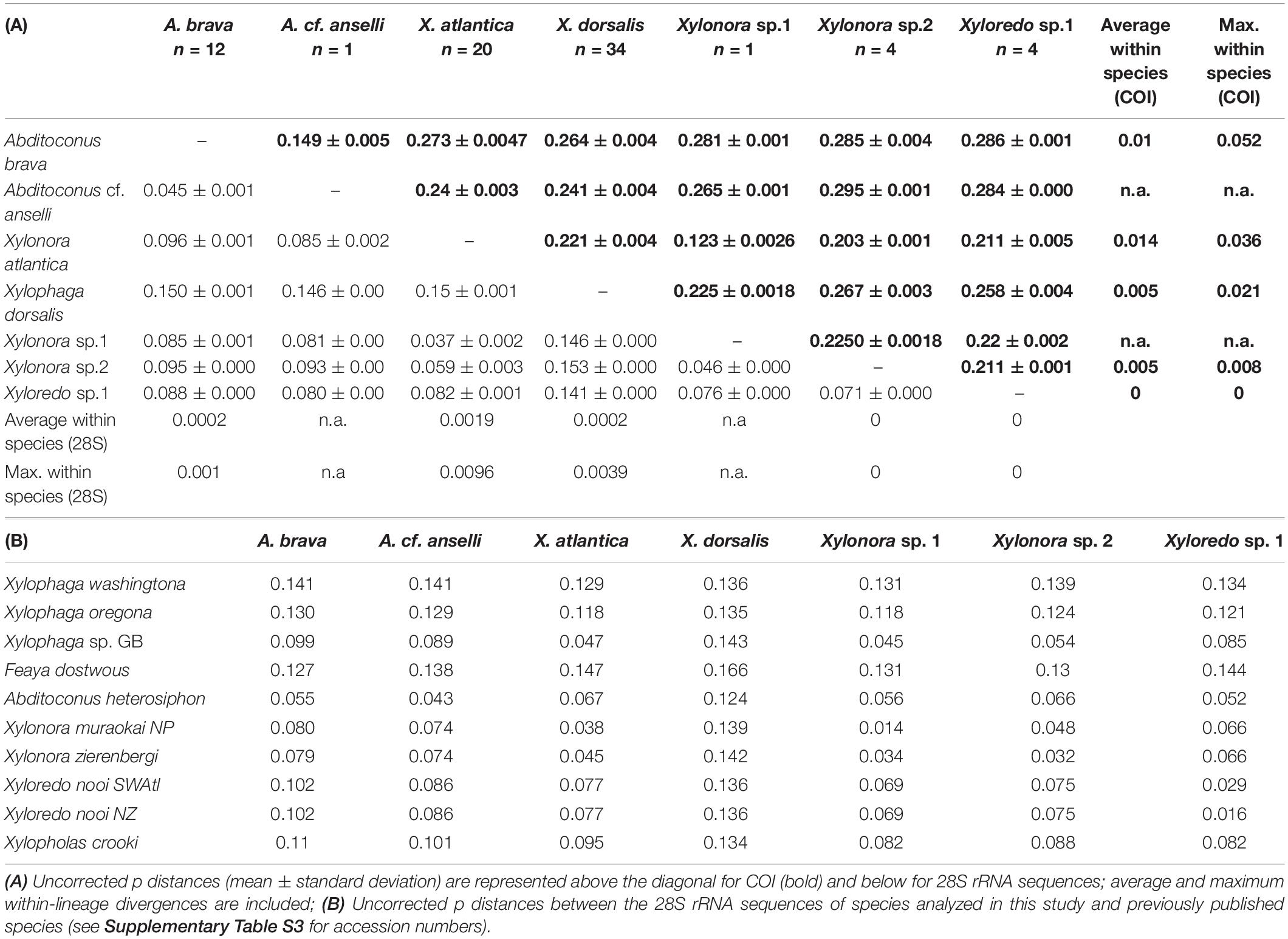
The COI analysis of the specimens considered in this study identified five sequence groups and two individual sequences (Supplementary Figure S1). They were all well separated from each other by uncorrected p-distances of ≥12.3% (COI) and 3.7% (28S rRNA), while distances within groups did not exceed 5.2% (COI) and 0.1% (28S rRNA), respectively (Table 3A). This suggested all seven represented separate species. These species groups were congruent with those in phylogenies recovered with concatenated 18S and 28S rRNA gene sequences (data not shown) and also matched morphological specimen determinations.
In the phylogenetic analysis of concatenated COI, 18S and 28S rRNA, all 32 individuals of X. dorsalis formed a well-defined sister lineage to X. oregona/X. washingtona within the major clade 3 (Figure 3). X. atlantica new comb. (21 individuals) and Xylonora sp. 1 (1 individual; clade 4) displayed the closest relationship among all recovered species groups (12.3% uncorrected p for COI; Table 3A). Their separation was supported by ML/BI analyses of the COI (75% bs, 0.999 pp; Supplementary Figure S1), while ML did not reproduce support in the analyses of concatenated genes (Figure 3). Because of the genetic distance between these two taxa and consistent grouping by ML and BI with different genes, we consider them as separate species. Xylonora sp. 2 was well separated from all other species, revealing a close relationship with X. atlantica new comb. (uncorrected p: COI 20.3%, 28S rRNA 5.9%; Table 3A) and Xylonora zierenbergi new comb. (28S rRNA 3.2%, Table 3) which is affiliated with high support to clade 4 (Figure 3). A. brava and the single sequence of Abditoconus cf. anselli represented well supported (Figure 3) and closely related sister lineages within clade 5 (uncorrected p: COI 14.9%, 28S rRNA 4.5%). Xyloredo sp. 1 was well separated from all other species in this study (uncorrected p: COI ≥ 21.1%, 28S rRNA ≥ 7.1%). Its position within Xyloredo (clade 7) was well supported by ML and BI analyses and this position was also confirmed by the close relationship to Xyloredo nooi from New Zealand (28S rRNA uncorrelated–p distance 1.6; COI sequence not available for X. nooi). The 28S rRNA uncorrelated-p distance between Xyloredo sp. 1 and X. nooi exceeded significantly any value we measured within any other species (max. 0.1). Xyloredo sp. 1 and X. nooi therefore appear to represent two separate species.
Morphological Observations
Our specimens of X. dorsalis, X. atlantica new comb. and A. brava match the original descriptions and additional observations are highlighting the intraspecific variability of their mesoplax and siphons (Romano et al., 2014). The specimens of Abditoconus cf anselli from the Avilés Canyon matched to certain degree the description of the type material from the Hebridean Slope (NW Scotland) and they clearly differed from A. brava (Romano et al., 2014). However, the original description of A. anselli was based on juveniles (<1 mm) and the description was generally poor. Thus the identity of our material cannot be formally confirmed. The specimens of Xylonora sp. 2 from Mid-Atlantic Ridge and Avilés Canyon (Figure 3) resemble the East Pacific Xylophaga microchira Voight, 2007 in shell shape and in having cirri at both siphon openings (Janet Voight, pers. obs.). However, Xylonora sp. 2 differs in having a small mesoplax with two plates joining at acute angle and subequal siphons, closely associated, shorter than shell length, the excurrent siphon without long and curved cirri and opening subterminally and dorsally to incurrent siphon. Larger individuals have small individuals attached dorsally (Figures 4E,F), but the taxonomic relevance of this character is doubtful. These dissimilarities, combined with the geographical separation, suggest that our specimens represent a different species. Unfortunately, no molecular data are available for the type specimens of Xylophaga microchira and further analyses are required to confirm the identity of our material.
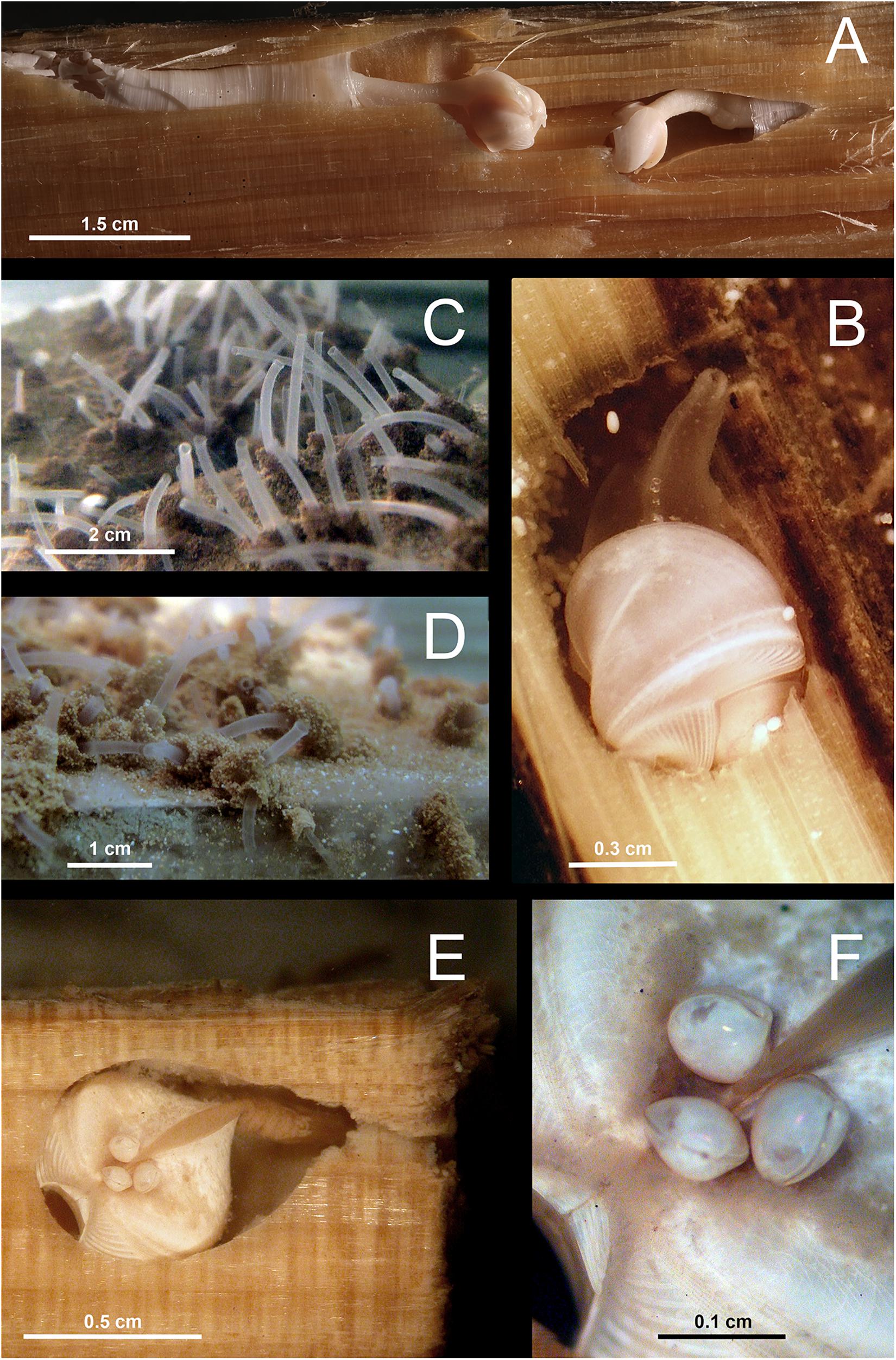
Figure 4. Examples of Xylophagaidae in experimentally deposited wood. (A) Xyloredo sp. 1 in pine wood from Avilés Canyon, 2,000 m depth. (B) Xylophaga dorsalis in wood from La Fonera Canyon, 130 m deep. (C,D) Living Xylophagaidae from La Fonera Canyon, 1100 m deep, kept in aquarium for several days after recovery. (C) Siphons emerging from wood and (D) fecal chimneys around the siphons. (E) Xylonora sp. 2 in a Chemecoli fir wood cube from the Rainbow hydrothermal vent field with small individuals attached on the dorsal shell posterior to umbo. (F) Close-up of the small individuals attached to Xylonora sp. 2.
The morphology of the single juvenile of Xylonora sp. 1 found in Avilés Canyon largely resembled X. atlantica new comb., but the separation from that species was supported by the presence of two small elongated and curved mesoplax plates resembling those of X. gagei instead of the triangular plates present in X. atlantica new comb. However, its small size (0.6 mm) and the fact that siphons were destroyed during sample handling actually prevented a consistent morphological assignation.
Xyloredo sp. 1 (Figure 4A) had calcareous tubes lining the burrows and differed in the shapes of shells and posterior adductor muscle scars from the previously known Atlantic species Xyloredo nooi and Xyloredo ingolfia Turner, 1972. However, its correct identification would require comparison with type material of all known species, which is beyond the scope of this paper.
Geographic Distribution and Intraspecific Genetic Structure of the COI Gene
We recovered seven xylophagaid species. Four of them occurred exclusively in the Atlantic (Abditoconus cf. anselli, Xylonora sp. 1, Xylonora sp. 2 and Xyloredo sp. 1), while no species was restricted to the Mediterranean.
Xylophaga dorsalis (Figures 4B, 5) showed the widest geographic distribution, being present in all locations in the Mediterranean and Atlantic Ocean, except for the northernmost Haakon Mosby Mud Volcano. Its distribution covered a wide bathymetric range from 2,000 m depth to our shallowest sites (Morocco mud volcanoes: 350 m depth; Western Mediterranean canyons: 130–550 m depth; Figures 1, 3 and Table 1) and a temperature range of 4 to 14°C (Table 1).
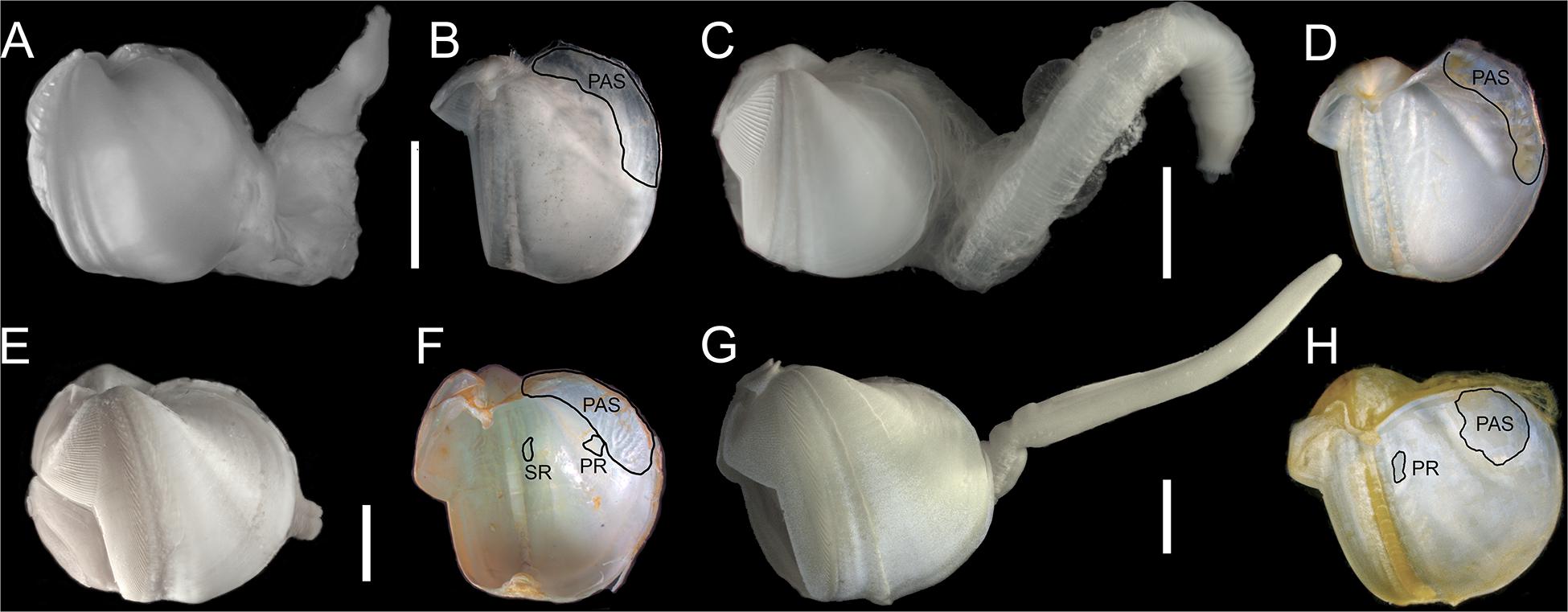
Figure 5. Lateral views of shell with soft parts and inner shells: (A,B) Abditoconus cf. anselli; (C,D) Abditoconus brava; (E,F) Xylonora atlantica new comb.; (G,H) Xylophaga dorsalis. PAS = posterior adductor scar; PR = pedal retractor scar; SR = siphonal retractor scar. Redrawn from Romano et al. (2014).
The COI sequence analysis indicated genetic connectivity across a large geographic range from Mid-Atlantic Ridge, Morocco Mud Volcanoes and throughout the Mediterranean. 29 individuals covering all sampling sites yielded 10 haplotypes (16 segregating sites, maximum within-clade uncorrected p distance: 2.1%). The dominant haplotype was shared by 62.1% of the individuals and it occurred in all sites except for Avilés Canyon. At this site we analyzed a single individual which diverged from the major haplotype by a single substitution (Figure 6). Half of all other haplotypes which were all collected in the Mediterranean were similarly closely related.
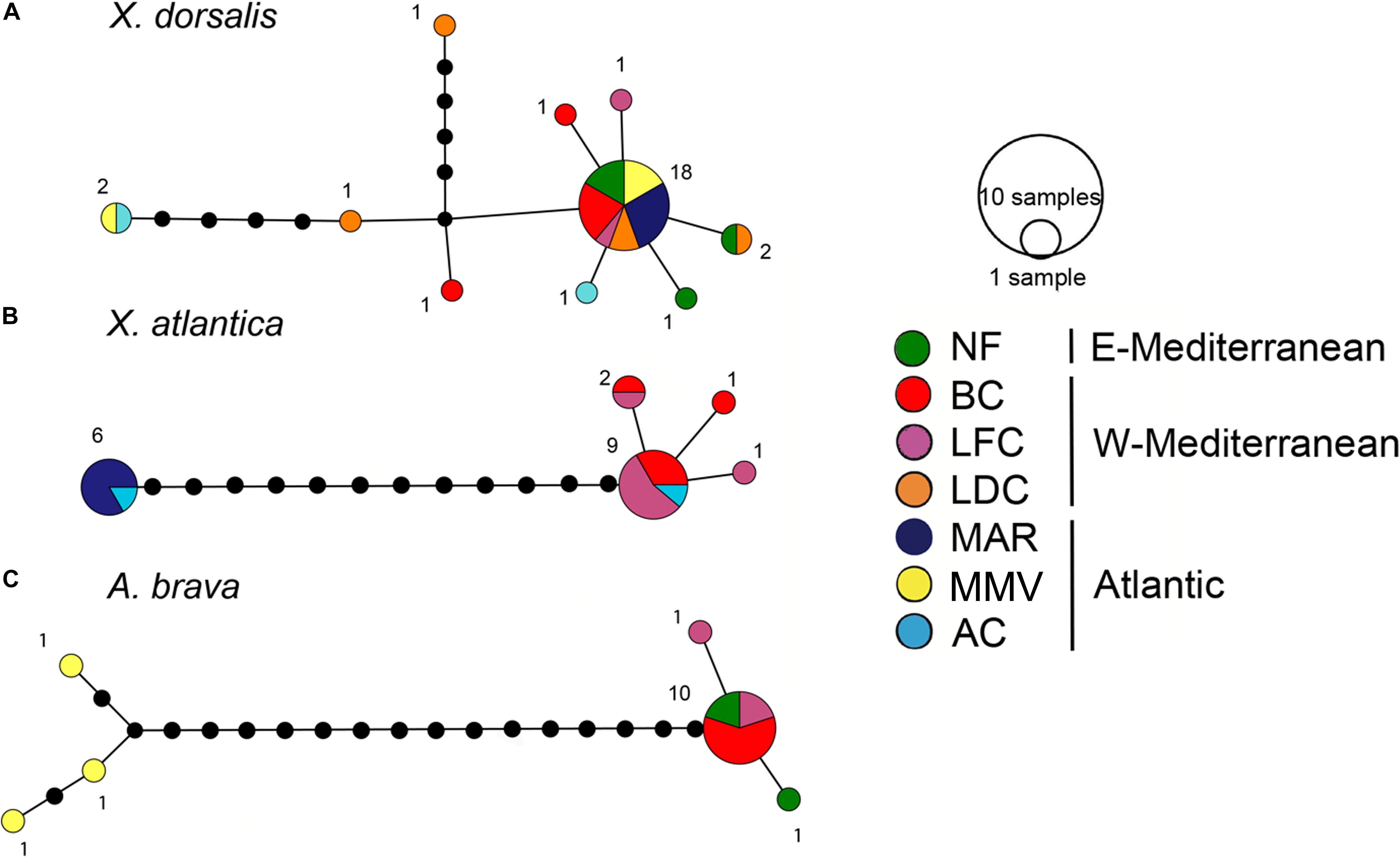
Figure 6. COI haplotype networks. (A) Xylophaga dorsalis (388 bp, 29 individuals). (B) Xylonora atlantica new comb. (388 bp, 19 individuals). (C) Abditoconus brava (389 bp, 15 individuals). Location colors as in Figure 1, except for MAR = Menez-Gwen hydrothermal vent filed at Mid-Atlantic Ridge, for location abbreviations see Figure 1.
Xylonora atlantica occurred in the temperate Atlantic Ocean (Mid-Atlantic Ridge and Avilés Canyon) and in the western Mediterranean, while it was not retrieved from Morocco mud volcanoes and the Eastern Mediterranean (Figures 1, 5). Its distribution covered an intermediate bathymetric range (870–2,000 m depth), with temperatures ranging from 4 to 13°C. The COI haplotype analysis of 19 sequences revealed five haplotypes (S = 16) displaying intraspecific divergence between the Mediterranean and Atlantic populations (Figure 6B). The dominant haplotype (47.4%) was present in Avilés Canyon and in the Western Mediterranean, but not on the Mid-Atlantic Ridge (Figure 6B). The second dominant haplotype (31.6%, within-clade divergence ≤3.6%) diverged by 13 substitutions from the dominant one and was exclusively recovered from the Atlantic, at Mid-Atlantic Ridge and Avilés Canyon (Figure 1). Three satellites of the dominant haplotype (1 substitution) occurred in four individuals from Western Mediterranean (Figure 6B).
Abditoconus brava was recovered from Eastern and Western Mediterranean and from the adjacent Morocco mud volcanoes, but not from any other Atlantic location (Figures 1, 5). The depth range was 700–1,700 m (Figure 3) and all collection sites shared the same habitat temperature of 13°C. The COI analysis (15 individuals) showed a clear separation between Mediterranean and Atlantic haplotypes, suggesting limited gene flow across the Gibraltar Strait. Among the six haplotypes recovered (S = 23), three occurred exclusively in the Mediterranean, while the other three were retrieved from Morocco mud volcanoes (Figure 6C). The two haplotype groups were well separated (17–19 substitutions). The largest intraspecific divergence occurred between the two haplotypes from the most distantly separated sites (Morocco mud volcanoes and Eastern Mediterranean, 5.15%). The most dominant haplotype (66.7%) was collected in Western and Eastern Mediterranean suggesting genetic connectivity throughout this oceanic basin.
Two species were recovered from two collection sites. Xylonora sp. 2 was found on the Mid-Atlantic Ridge (2,300 m depth, 3.5°C) and in the Avilés Canyon (2,000 m depth, 4–9°C). Three COI haplotypes recovered from four individuals of Xylonora sp. 2 were closely related to each other (S = 3, ≤2 substitutions) and different haplotypes occurred in the two collections sites. Xyloredo sp. 1 occurred in the Avilés Canyon in the Bay of Biscay (1200–2000 m depth, 4–9°C) and at the 3,300 km distant Haakon Mosby Mud Volcano (1240 m depth, at −1°C; COI sequences only available from this site). Two other species, Abditoconus cf. anselli (Figure 5) and Xylonora sp. 1, were exclusively recovered from the Avilés Canyon (2,000 and 1,200 m deep, respectively, 4–9°C).
Discussion
This study provides the first DNA barcode approach to identifying Xylophagaidae, where we combined molecular data (based on COI, 18S and 28S rRNA sequences) with morphology to discriminate between species and to assess their geographic distributions. Our analyses revealed: (i) a reliable species discrimination based on COI; (ii) a robust sequence divergence supporting the existence of seven species, three of them unidentified (two morphologically similar to X. atlantica new comb. and a possible undescribed Xyloredo); (iii) wide genetic connectivity throughout the North Atlantic and the Mediterranean for Xylophaga dorsalis, the most abundant species in our samples, and for Xylonora atlantica new comb.; and (iv) limited genetic connectivity between the Atlantic and Mediterranean populations of A. brava.
Phylogeny of Xylophagaidae
Our results (based on 18S rRNA, 28S rRNA and COI genes) agree with those based on 18S and 28S rRNA by Distel et al. (2011) in showing the Teredinidae and Xylophagaidae as sister taxa sharing a xylotrophic common ancestor. Reports considering xylophagaids as a family referred to the names Xylophaginidae, Xylophagidae, and Xylophagaidae (Purchon, 1941; Knudsen, 1961). In order to avoid homonymy with a taxon of Diptera [Xylophagide Fallén (1810), Insecta], the name was amended to Xylophagaidae (International Commission on Zoological Nomenclature, 2018) that has been used most recently (Gaudron et al., 2010; Coan and Valentich-Scott, 2012; Haga and Kase, 2013; Romano et al., 2014; Rosenberg and Gofas, 2015; Voight et al., 2019).
Our phylogenetic reconstruction (Figure 3) revealed that Xylophaga is polyphyletic, unless the new genus Xylonora is erected for the species in clade 4. Our decision to separate it from Xylophaga (clade 3) is supported by robust morphological and taxonomic arguments. The rapid evolutionary substitution rate of the mitochondrial COI, compared to the nuclear 18S and 28S rRNA genes, allows for discriminating closely allied species and also different phylogeographic groups within species. For the same reasons, its phylogenetic signal is weak (Hebert et al., 2003; Cunha et al., 2009). However, a number of studies on bivalve phylogenies proved that combining nuclear and COI markers improved the signal (Giribet and Distel, 2003; Bieler et al., 2014; Johnson et al., 2017). Our data support the monophyly of Xylophaga sensu stricto and Xylopholas. This is in contrast to previous results that were based on 18S and 28S rRNA analyses, but did not include COI (Voight et al., 2019). Additional support for the separation of Xylonora new genus and Xylophaga sensu stricto may come from recent observations on different trophic ecology of the two genera (Voight et al., 2020). Three members of the Xylophaga sensu stricto clade (X. dorsalis, X. washingtona, and X. alexisi) presented higher δ15N signatures than other Xylonora-clade species in which δ15N values were consistent with strict xylotrophy. This may suggest that Xylophaga sensu stricto can filter feed as an opportunistic alternative to xylotrophy, while members of Xylonora are strict xylotrophs (Voight et al., 2020). Our data are furthermore decisive enough to confirm the sister-taxa relationship between representatives of Xylophaga in the Atlantic (X. dorsalis) and the Pacific (X. washingtona/X. oregona).
Species Identification Based on Molecular Markers
DNA barcoding allowed us to improve taxonomic resolution to assess the species richness and diversity within xylophagaids. As in shipworms, the high intraspecific phenotypic plasticity of xylophagaids is often not well documented and it makes identifications difficult. Shell morphology often appears to vary in relation to environmental conditions. For instance, shell shape in X. washingtona recovered from wood deployments at 2,370 m depth off California varied in relation to different wood substrates (i.e., cedar, pine, ash, maple, oak). The variations included valve size and number of denticulated ridges on the anterior slope in relation to valve length (Turner, 2002). Xylonora atlantica new comb. from wood deployments off Cape Cod (MA, United States) revealed different growth rates and a morphology varying during the year as well as with site, substrate, and animals’ crowding on wood (Romey et al., 1994). Xylophaga dorsalis and Xylonora atlantica new comb. recovered from pine and oak in the submarine canyons of Avilés (Bay of Biscay), Blanes and Lacaze-Duthiers (Western Mediterranean) also showed phenotypic variability in a previous study (Romano et al., 2014). Our materials of X. dorsalis, X. atlantica new comb, and A. brava exhibited comparable phenotypic variations among conspecifics collected from pine and oak in an even wider geographic and environmental range in the Atlantic and Mediterranean (Table 1). However, ambiguities were well-resolved by the three molecular markers used, which all showed clear and identical clustering (Figure 3).
Interspecific disparity can be characterized with thresholds of genetic distance between sequences, which may vary according to the compared taxa and the molecular markers. In COI of birds, an interspecific genetic distance of ten times the average distance within species was suggested (Hebert et al., 2004), while in marine gastropods 4.9–7.8 times revealed to be conservative enough to eliminate false positive species discrimination (Meyer and Paulay, 2005). The wide range of species attributes among these gastropods (e.g., fertilization modes, larval dispersal and feeding strategies) allowed for suggesting that these findings should be applicable to a wide range of marine taxa (Meyer and Paulay, 2005).
The 28S rRNA and COI genetic distances clearly characterized species disparity in our materials based on the “ten times-threshold” criterion. Both markers showed clear barcoding gaps between species without overlapping ranges of intraspecific and interspecific distances (Meyer and Paulay, 2005) and the average distance between two species was as a rule >10 times larger than the average distance within each species (Table 1). The only exception occurred in the comparison of the single specimen of Xylonora sp. 1 (initially misidentified as X. atlantica) and Xylonora atlantica new comb., which revealed an interspecific distance of only 8.2× the distance within X. atlantica new comb. However, this was well beyond the 4.9–7.8 threshold derived from marine gastropods and we are very confident in that they are two separate species. Xylonora sp. 2 and Xyloredo sp. 1 are also well separated from the other species within the respective genera. Overall, more individuals of these three morphotypes are needed to allow precise morphological identifications or formal new species descriptions.
Diversity of Xylophagaidae in European Deep Waters and Ecological Considerations
Thirteen xylophagaid species are currently recognized from the North-Eastern Atlantic and adjacent marginal seas. Apart from the seven species covered by this study, there are Xylophaga praestans, Xylophaga nidarosiensis Xylophaga noradi, Xylophaga ricei, Xylophaga gagei and Xyloredo ingolfia. Xylophagaid diversity in European waters thus exceeds the diversity of teredinids in the same area that consists of nine species (Borges et al., 2014). The higher diversity of Xylophagaidae is remarkable with regard to the, at first glance, very similar habitat requirements and feeding ecologies of the two families and also because abundance and variety of wood are assumed much less in the deep sea than in coastal-near shallow regions. This difference in species diversity cannot be explained by an artifact from sampling bias because xylophagaids from the deep-sea floor are difficult to access and have been investigated far less than shallow water Teredinidae. We hypothesize that factors such as differential adaptation to depth, temperature and salinity and related water mass chemistry are leading to habitat partitioning, which may affect species diversity differently in the two families.
While teredinids are bound to shallow waters, most European xylophagaids cover either very broad bathymetric ranges from the littoral to the abyss (e.g., X. atlantica new comb. and X. dorsalis) or were exclusively recovered from bathyal or abyssal depths (Table 4). Only Xylophaga nidarosiensis and Xylophaga noradi were solely recovered from shallow waters. However, both species are only known from single records and our view on their bathymetric distributions may change with future collections.
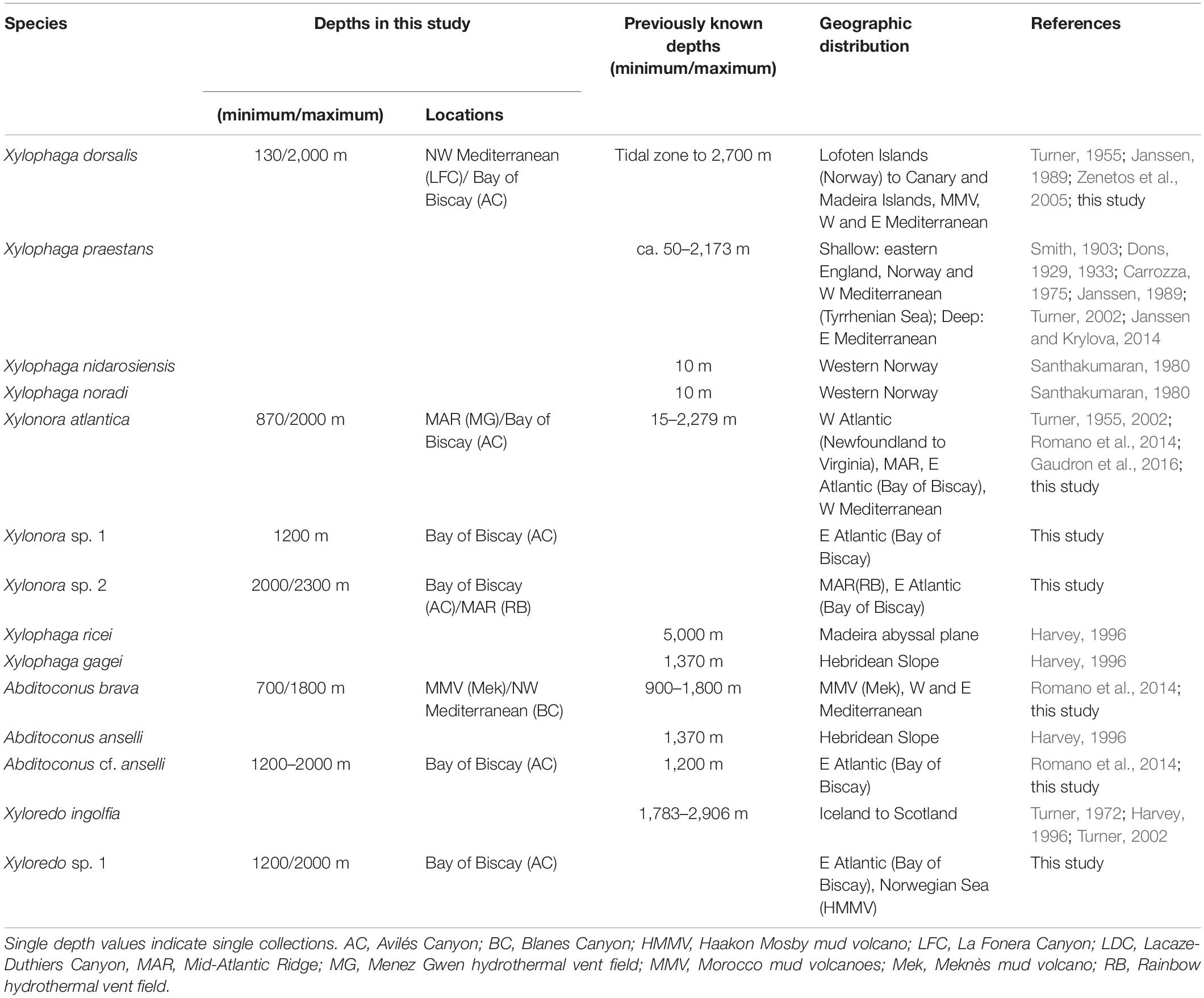
Table 4. Depth distributions (minimum/maximum) of the xylophagaid species recorded in the North East Atlantic and adjacent marginal seas.
Shallow water species from temperate oceans are generally eurythermic because they must cope with seasonally changing water temperatures. This applies to teredinids and also some xylophagaids. Eurythermic xylophagaids additionally covering broad depth ranges in the north-eastern Atlantic must also tolerate constant low temperatures. Examples are Xylonora atlantica new comb. and Xylophaga dorsalis which can tolerate 23 and 20°C, respectively (Borges et al., 2014), and they also live in bathyal to abyssal depths where temperatures are constantly low (Table 4). Other species may be cold-stenothermic, e.g., Xyloredo sp. 1 which we recovered at −1 to +4°C, or Xyloredo ingolfia, which occurs only in the deep northern Atlantic. Most other European xylophagaids may be also cold-stenothermic with regard to the depths from which they were retrieved, however, only sporadic or single collections of many xylophagaid species prevent clear interpretations of their temperature tolerances.
North East Atlantic mid- and deep-water fauna is adapted to a salinity range of 34.4–35.3‰ (Bouchet and Taviani, 1992; Emery, 2001). Atlantic xylophagaids living exclusively below 500 m depth may therefore be typical stenohaline deep-sea organisms. Species living in shallower Atlantic waters must tolerate higher salinities of 35.2–36.7‰ (Emery, 2001) and species living in the deep Mediterranean must cope with >38.0‰ (Miller et al., 1970). Xylophagaids covering broad salinity ranges in the Atlantic and occurring also in the Mediterranean such as Xylonora atlantica new comb., Xylophaga dorsalis and Abditoconus brava are therefore considered euryhaline marine species (Table 4).
Species diversity estimations can be affected by factors that influence seasonal colonization times such as variations of local current regimes or different reproduction and spawning times in different species. Temporal effects became apparent in the eastern Mediterranean because X. dorsalis was the only colonizer in 2007 while A. brava appeared only in 2009 and became the dominant species. Different colonizers may also be able to select different types of wood. A. brava for example was almost exclusively found in pine and not in oak when both types of wood were simultaneously deployed in the Western Mediterranean canyons.
Our study counteracted the risks of ignoring seasonally occurring species by repeated sampling in most sites (Table 1). Among them, the highest diversity of six species occurred in Avilés Canyon (Figure 1). This site provided various depth and temperature conditions of 1,200 to 2,000 m and 4 and 9°C, respectively, and thus it certainly combined suitable conditions for the generalists X. atlantica new comb. and X. dorsalis and also for putatively cold-stenothermic/stenohaline deep-water species such as Abditoconus cf. anselli, Xylonora sp. 1, Xylonora sp. 2, Xyloredo sp. 1. Furthermore, the close vicinity of Avilés Canyon to the coast should support a regular and sufficient reception of wood allowing for continuous maintenance of high species diversity. Our Eastern Mediterranean site showed low diversity of only two species suggesting that only eurythermic and euryhaline species like X. dorsalis and A. brava were able to tolerate the high salinity and temperature of 13°C at 1200 water depth (Miller et al., 1970).
Dispersal Capabilities
Our results prove that xylophagaids occur in the entire North East Atlantic, from the Barents Sea in the Arctic, along the northern European continental margin and over the temperate Mid-Atlantic Ridge, to the far Eastern Mediterranean. This is a remarkable distribution range if we assume that land-derived wood is only scarcely available in remote areas such as Arctic deep waters or the Mid-Atlantic Ridge.
Colonization of experimentally deployed wood proves that xylophagaids disperse in the deep ocean and not by colonizing driftwood before it sinks down. The colonization of wood in remote oceanic regions is a strong indication for high dispersal capabilities and for bridging large distances by xylophagaid larvae.
Deep-sea species with long-distance dispersal are usually assumed to have planktotrophic larvae, particularly when they inhabit fragmented and widely separated habitats (Tyler and Young, 1999). Overall knowledge on xylophagaids’ larval biology remains scant. Planktotrophic development has been documented at least for the Atlantic X. atlantica new comb. and the Western Pacific Xylophaga supplicata (Taki and Habe, 1950), both exhibiting typical shell morphologies of long-lived pelagic larvae (Gaudron et al., 2010; Haga and Kase, 2013). Laboratory experiments with reared larvae of X. atlantica new comb. showed that metamorphosis can be delayed for up to 6 months (Culliney and Turner, 1976), mirroring the larval trophic strategies, planktonic larval duration and dispersal capacities of other deep-sea bivalves such as the hydrothermal vent and cold seep subfamily Bathymodiolinae. They also live in fragmented habitats, sometimes separated by hundreds of kilometers, and disperse with planktotrophic larvae that can remain up to 13 months in the plankton (Arellano and Young, 2009). We may thus reasonably assume that xylophagaids other than X. atlantica new comb. and X. supplicata may also have equivalent dispersal strategies.
Biogeography of European Xylophagaidae
All thirteen European xylophagaids occurred in the temperate North East Atlantic, while only four occurred also in the Mediterranean (Xylophaga dorsalis, Xylophaga praestans, Xylonora atlantica new comb., Abditoconus brava). Compared to the nearby Atlantic, the Mediterranean deep sea generally harbors an impoverished fauna and cold stenothermic species are lacking (Fredj and Laubier, 1985; Breusing et al., 2016). This appears to result from the isolation of the Mediterranean and its desiccation during the Messinian Salinity Crisis in the late Miocene (Cita, 1973; Hsü et al., 1973), followed by its recolonization only after reconnection with the Atlantic (Pérès, 1985; Bouchet and Taviani, 1992). Since then, the Gibraltar Strait and the Siculo-Tunisian sills in combination with the significantly different hydrological conditions supposedly acted as biogeographic barriers controlling the Atlanto–Mediterranean benthic exchanges and preventing establishment of cold-stenothermic and stenohaline deep-sea fauna in the Mediterranean (Bouchet and Taviani, 1992). All xylophagaids occurring in the Mediterranean are considered eurythermic and euryhaline and it is reasonable to assume an Atlantic origin.
Xylophaga dorsalis has a wide distribution range in the East Atlantic, from the Lofoten Islands in the northern Norwegian Sea and the British Isles to the Canary and Madeira Islands and the Western and Eastern Mediterranean (Turner, 1955; Romano et al., 2013, 2014; this study). We recorded this species for the first time on the Mid-Atlantic Ridge. Its bathymetric range covers littoral to abyssal depths (Table 4) and it is considered eurybathic, eurythermic and euryhaline. It was also the most common and abundant species in our study sites.
Xylophaga praestans was recorded in shallow northern European waters (British Isles to Norway) and in the Mediterranean down to bathyal depths (Table 4). There are doubts on its previous identification and depth range in the deep Eastern Mediterranean (Mienis, 2003). Re-examination of this material is beyond the scope of this study, and we therefore preliminarily consider this species as eurybathic, eurythermal, and euryhaline.
Xylonora atlantica new. comb. is a pan-Atlantic species, well known from the North West Atlantic margin (Turner, 1955; Culliney and Turner, 1976; Romey, 1991; Romey et al., 1994; Voight, 2008; Distel et al., 2011) and only recently recovered from the south European Atlantic margin and the Western Mediterranean (Romano et al., 2014). We found this species also on the Mid-Atlantic Ridge, while it appears to be absent from the Eastern Mediterranean and northern European waters. Its bathymetric distribution ranges from littoral to abyssal depths (Table 4) and it is therefore considered eurythermic and euryhaline.
We recovered Xylonora sp. 2 from bathyal depths in two locations, the Avilés Canyon and the Mid-Atlantic Ridge and we therefore consider it as a stenothermic and stenohaline deep-sea species.
Abditoconus brava was previously known from the Western Mediterranean (Romano et al., 2014). We recorded it also in the adjacent Atlantic (Morocco mud volcanoes) and in the Eastern Mediterranean. A. brava appears to be a bathyal species (Table 4) tolerating also the high temperatures and salinities in the deep Eastern Mediterranean.
We found Xyloredo sp. 1 in Avilés Canyon and the Haakon Mosby Mud Volcano in the Arctic which represents the northernmost record for a Xylophagaidae. This species occurs in bathyal depths and it is considered cold-stenothermic and stenohaline.
Xyloredo ingolfia is a bathyal species occurring from Iceland to Scotland in the North East Atlantic (Turner, 1972, 2002; Harvey, 1996) and it is therefore considered cold-stenothermic and stenohaline.
All other European xylophagaids were recovered only once, all of them from littoral to abyssal depths at different locations among Norway, the British Isles and Avilés Canyon (Table 4).
Inter-Population Connectivity
Our discovery of identical COI haplotypes among the populations of X. dorsalis, X. atlantica new comb., and A. brava sampled in distant locations in the Atlantic and the Mediterranean strongly suggest gene flow over large distances.
The presence of an identical major COI haplotype in X. dorsalis across most of our study sites indicated unhindered genetic connectivity between the Mid-Atlantic Ridge and Morocco mud volcanoes, across the Gibraltar Straight and throughout the entire Mediterranean. This is in accordance with the above hypothesized eurybathic, eurythermic and euryhaline nature of this species which also accounts for tolerance of their larvae toward contrasting environmental conditions in shallow and deep environments of the temperate Atlantic and the warm Mediterranean. The single haplotype retrieved from Avilés Canyon in the Bay of Biscay diverged by only one substitution, and it is likely that more extensive sampling at this site would have revealed the major haplotype as well. We may therefore reasonably assume unimpeded genetic exchange among the populations of X. dorsalis in an area exceeding the coverage of our study, within its entire distribution range between northern Europe and the Canary Islands.
Genetic connectivity between the Atlantic and the western Mediterranean was also evidenced for eurybathic and eurythermic X. atlantica new comb. Its major haplotype was sampled in Avilés Canyon and in the Western Mediterranean submarine canyons. One divergent haplotype was recovered exclusively in the Atlantic on the Mid-Atlantic Ridge and the Avilés Canyon (Figure 6). Future research must reveal if this haplotype is probably representing a typical one for western Atlantic populations and if this may indicate limited connectivity between the Atlantic and the Mediterranean populations.
In A. brava, the presence of an identical major haplotype in the Western and Eastern Mediterranean populations suggests unobstructed gene flow throughout the entire Mediterranean. This haplotype and a few closely related ones were limited to the Mediterranean, while a clearly divergent haplotype was recovered exclusively at Morocco mud volcanoes in the adjacent Atlantic. We cannot discard that our limited sampling effort caused a bias and that future research may recover the “Atlantic haplotype group” in the Mediterranean and vice versa. However, the 16-substitution divergence strikingly exceeded what we observed in Xylonora atlantica new comb. and Xylophaga dorsalis across the Atlantic-Mediterranean transition. A. brava was not recovered from shallower than 700 m depth, and it is possibly a strictly bathyal species. The less than 300-m deep Gibraltar Strait at the entrance of the Mediterranean (Gascard and Richez, 1985) could then represent a real biogeographic barrier, significantly limiting gene flow. If so, we may assume a continuing divergence in the Atlantic and Mediterranean populations, which may eventually lead to speciation.
Conclusion
Our study integrates morphological and molecular data, including DNA barcoding of deep-sea wood boring bivalves. We thus provide a molecular baseline that is significantly facilitating species discrimination and will greatly enhance taxonomic knowledge in the future. Molecular data confirmed the Xylophagaidae as being composed of seven well-supported major clades corresponding to independent taxa with clear morphological differences. This leads us to erect Xylonora as a new genus to resolve the polyphyly of Xylophaga.
Repeated recoveries of Xylophagaidae from wood deployed experimentally over wide geographic, bathymetric, temperature and salinity ranges gave insights into the biogeography of European species for the first time, allowing for conclusions on the ecological requirements reaching far beyond what can be drawn from single records. We recovered seven species, some of them occurring regularly (e.g., Xylophaga dorsalis and Xylonora atlantica new comb.), showing wide Atlanto-Mediterranean distributions, broad bathymetric, temperature and salinity ranges, and genetic connectivity among their widely separated populations. This allowed us to conclude that a broad tolerance toward different hydrographic regimes may be a major clue for their dispersal success.
The biogeographic patterns of other Xylophagaidae may be linked to lower tolerance toward shallow water environments or temperature and salinity variations. Xyloredo sp. 1, for example, was exclusively collected in bathyal sites in the temperate North Atlantic, while A. brava occurred in bathyal regions throughout the Mediterranean and nearby Atlantic. For this species, the shallow Gibraltar Strait is likely representing a biogeographic barrier limiting gene flow.
The presence of xylophagaids in remote deep-sea locations such as Haakon Mosby Mud Volcano in the Arctic or the Mid-Atlantic Ridge where wood is presumably a scarce habitat suggests that distance alone is not a limiting factor for their dispersal. Rising frequencies of major storms and related flooding of the coasts caused by changing climate conditions may probably enhance the future availability of wood as stepping stones for the dispersal of wood-colonizing benthic organisms on the deep-sea floor. This may also affect the distribution of other wood colonizing deep-sea fauna.
Our results prove that experimentally deployed wood-falls could be used as efficient tools to collect wood specialists, highlighting their potential in future deep-sea studies addressed to improve our knowledge on their population dispersal abilities and connectivity. This may certainly include also macrofaunal taxa not specialized to wood and such research could provide crucial knowledge for planning strategies for a sustainable management of deep-sea species and ecosystems.
Data Availability Statement
All new sequences were deposited in NCBI under Bioproject PRNEB39839; accessions for target genes are COI: LR899288-LR899358, 18S rRNA: LR899359-LR899420, and 28S rRNA: LR899226-LR899286. All accession numbers used in this study can be found in the Supplementary Material.
Author Contributions
CR, AN-J, DM, NL, and CB conceived and designed the experiments. CR, AN-J, CB, and NL performed the experiments. CR, AN-J, CB, and GR analyzed the data. CR contributed to taxonomic identifications. CR, AN-J, DM, and CB wrote the final manuscript. All authors wrote the draft manuscript.
Funding
This work was supported by the Max Planck Society and the French Centre National de la Recherche Scientifique (CNRS) associating Université de Liège, UPMC, and Museum of Natural History through the GDRE program “Diversity, establishment and function of organisms associated with marine wood falls-DiWOOD”; by project CHEMECO (European Sciences Foundation (ESF)/Eurocores/EURODEEP/0001/2007); by the Chair programme “Extreme Marine Environments, Biodiversity and Global Change” UPMC-Fondation TOTAL; by the EUROFLEET Programme; by the Agencia Española de Investigación (AEI) and the European Funds for Regional Development (FEDER/UE) through the research projects PROMETEO (CTM2007-66316-C02-02/MAR), DOSMARES (CTM2010-21810-C03-03) and PopCOmics (CTM2017-88080); and by Agència de Gestió d’Ajuts Universitaris i de Recerca of the Generalitat of Catalunya through the Consolidated Research Group on Marine Benthic Ecology (2017SGR378). Shiptime during research cruises BioBaz, M70/2-Bionil, MSM13/3-Homer, ARKXXII/1b, ARKXXIV/2, Medeco-2, and POS403-MenezKart received funding from the EU 6th FP HERMES (GOCE-CT-2005-511234), EU 7th FP HERMIONE (grant agreement no. 226354), CNRS, and the DFG. CR was funded by the People Programme (Marie Curie Action IOF to CR) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under the “DeepFall” project http://www.deepfall-project.eu; https://twitter.com/DeepFall_Proj (REA grant agreement N. PIOF-GA-2013-628146); AN-J was funded through DiWOOD; CB was funded by the DFG Cluster of Excellence “The Ocean in the Earth System” at MARUM (University of Bremen), a European Research Council Advanced Grant (BathyBiome, Grant 340535) and the Max Planck Society.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the chief scientists, captains and crews and the teams of ROV Victor 6000 (IFREMER, France), ROVs MARUM Quest 4000 and Cherokee (MARUM, Bremen, Germany), ROV Isis (NOC Southampton, United Kingdom), and ROV Genesis (Renard Centre of Marine Geology, Belgium) for their help during the following cruises: DOSMARES (NW Mediterranean canyons) with the Spanish RV Garcia del Cid; BIOCANT (Avilés Canyon) with the Spanish RV Sarmiento de Gamboa; MEDECO (2007, Eastern Mediterranean), MoMARDream-Naut (2007) and MoMARDream-08 (2008, both to Rainbow hydrothermal vent field), (https://doi.org/10.17600/7030060) and BIOBAZ (2013, Menez Gwen hydrothermal vent field) with the French RV Pourquoi Pas? BIONIL/M70-2 (2006, Eastern Mediterranean) with the German RV Meteor; HOMER/MSM13-3 (2009, Eastern Mediterranean) with the German RV MARIA. S. MERIAN; ARKXXII/1b (2007) and ARKXXIV/2 (2009, both to Haakon Mosby mud volcano) with the German RV POLARSTERN; MENEZKART/POS402 (2010, Menez Gwen hydrothermal vent field) with the German RV POSEIDON; JC-10 (2007, Mercator mud volcano, N-Atlantic) with the British RRS James Cook; 64PE284 (2008, Meknès mud volcano, N-Atlantic) with the Dutch RV Pelagia; B09/14 (2009, Mercator and Meknès mud volcanoes) with the Belgian RV Belgica. Wood colonization experiments in the Lacaze-Duthiers Canyon were deployed with the RV Minibex (COMEX). Material from the Morocco mud volcanoes was collected within the Chemeco (Monitoring colonization in chemosynthetic ecosystems) from the European Science Foundation under grant EURODEEP/0001/2007 and FTC (Fundação para a Ciência e a Tecnologia) project; we thank Dr. Marina Cunha, University of Aveiro, Portugal and Sylvie Gaudron, Sorbonne Université, France, for providing specimens. We are grateful to Janet R. Voight from the Field Museum of Natural History, Chicago, IL, United States, and Takuma Haga from JAMSTEC, Kanagawam, Japan, for taxonomic advice. We also thank S. Wetzel, D. Cortese, L. Anducas Comas, G. Carreras for their laboratory help, particularly their perseverance with the PCR and sequencing of such a difficult material.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.579959/full#supplementary-material
Supplementary Figure S1 | Bayesian Inference analysis (10,000,000 generations, sampling interval 1000×) based on COI sequences (388 bp). Statistical support (BI posterior probabilities >0.85/RAxML 100× bootstraps) is indicated on the nodes. Taxa names include locations (see Figure 1 for abbreviations) and identity of the specimens (see Supplementary Table S3 for GenBank accession numbers). Sampling locations are also indicated by color code.
Supplementary Table 1 | List of primers. ∗ = PCR primer pairs; unmarked = internal primers. Annealing temperatures (Ta, °C) modified for amplification steps depending on enzymes (see Supplementary Table S2).
Supplementary Table 2 | List of enzyme polymerases and the PCR conditions used in this study.
Supplementary Table 3 | Identity of the specimens used for each genetic analysis, and GenBank sequences and accession numbers. ∗ = outgroup species for tree reconstruction.
Footnotes
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Arellano, S. M., and Young, C. M. (2009). Spawning, development, and the duration of larval life in a deep-sea cold-seep mussel. Biol. Bull. 216, 148–162. doi: 10.1086/BBLv216n2p149
Baco, A. R., Smith, C. R., Peek, A. S., Roderick, G. K., and Vrijenhoek, R. C. (1999). The phylogenetic relationships of whale-fall vesicomyid clams based on mitochondrial COI DNA sequences. Mar. Ecol. Progr. Ser. 182, 137–147. doi: 10.3354/meps182137
Bartsch, P. (1921). A new classification of the shipworms and descriptions of some new wood boring mollusks. Proc. Biol. Soc. Washington 34, 25–32.
Bessette, S., Fagervold, S., Romano, C., Martin, D., Le Bris, N., and Galand, P. E. (2014). Diversity of bacterial communities on sunken woods in the Mediterranean Sea. J. Mar. Sci. Technol. 22, 60–66.
Bieler, R., Mikkelsen, P. M., Collins, T. M., Glover, E. A., Gonzalez, V. L., Graf, D. L., et al. (2014). Investigating the bivalve tree of Life – an exemplar-based approach combining molecular and novel morphological characters. Invert. Syst. 28, 32–115. doi: 10.1071/is13010
Bienhold, C., Pop Ristova, P., Wenzhöfer, F., Dittmar, T., and Boetius, A. (2013). How deep-sea wood falls sustain chemosynthetic life. PLoS One 8:e53590. doi: 10.1371/journal.pone.0053590
Bienhold, C., Wenzhöfer, F., Le Bris, N., Ramette, A., and Boetius, A. (2008). Wood colonization experiments in the eastern mediterranean deep sea. Geophys. Res. Abstr. EGU 2008:09061.
Borges, L. M. S., Merckelbach, L. M., Sampaio, I., and Cragg, S. M. (2014). Diversity, environmental requirements, and biogeography of bivalve wood-borers (Teredinidae) in European coastal waters. Front. Zool 11:13. doi: 10.1186/1742-9994-11-13
Borges, L. M. S., Sivrikaya, H., Le Roux, A., Shipway, J. R., Cragg, S. M., and Costa, F. O. (2012). Investigating the taxonomy and systematics of marine wood borers (Bivalvia : Teredinidae) combining evidence from morphology, DNA barcodes and nuclear locus sequences. Invert. Syst. 26, 572–582. doi: 10.1071/IS12028
Bouchet, P., and Taviani, M. (1992). The Mediterranean deep-sea fauna – pseudopopulations of Atlantic species. Deep Sea Res. Part A Oceanogr. Res. Pap. 39, 169–184. doi: 10.1016/0198-0149(92)90103-z
Breusing, C., Biastoch, A., Drews, A., Metaxas, A., Jollivet, D., Vrijenhoek, R. C., et al. (2016). Biophysical and population genetic models predict the presence of “phantom” stepping stones connecting Mid-Atlantic Ridge vent ecosystems. Curr. Biol. 26, 2257–2267. doi: 10.1016/j.cub.2016.06.062
Carrozza, F. (1975). Microdoride di malacologia mediterranea (Contributo Primo). Conchiglie 11, 185–192.
Chernomor, O., Von Haeseler, A., and Minh, B. Q. (2016). Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 65, 997–1008. doi: 10.1093/sysbio/syw037
Cita, M. B. (1973). “Mediterranean evaporite: paleontological arguments for a deep-basin desiccation model,” in Messinian Events in the Mediterranean, ed. C. W. Drooger (Amsterdam: North-Holland Publishing Co.), 206–228.
Coan, E. V., and Valentich-Scott, P. (2012). Bivalve Seashells of Tropical West America. Marine Bivalve Mollusks From Baja California to Northern Peru. Santa Barbara: Santa Barbara Museum of Natural History.
Combosch, D. J., Collins, T. M., Glover, E. A., Graf, D. L., Harper, E. M., Healy, J. M., et al. (2017). A family-level tree of life for bivalves based on a Sanger-sequencing approach. Mol. Phylogenet. Evol. 107, 191–208. doi: 10.1016/j.ympev.2016.11.003
Conrad, T. A. (1846). Descriptions of new species of fossil and recent shells and corals. Proc. Acad. Nat. Sci. Phil. 3, 19–27.
Culliney, J. L. (1975). Comparative larval development of the shipworms Bankia gouldi and Teredo navalis. Mar. Biol. 29, 245–251. doi: 10.1007/BF00391850
Culliney, J. L., and Turner, R. D. (1976). Larval development of the deep-water wood boring bivalve. Xylophaga atlantica Richards (Mollusca, Bivalvia, Pholadidae). Ophelia 15, 149–161. doi: 10.1080/00785326.1976.10425455
Cunha, M. R., Matos, F. L., Génio, L., Hilário, A., Moura, C. J., Ravara, A., et al. (2013). Are organic falls bridging reduced environments in the deep sea? – Results from colonization experiments in the Gulf of Cádiz. PLoS One 8:e76688. doi: 10.1371/journal.pone.0076688
Cunha, R. L., Grande, C., and Zardoya, R. (2009). Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evol. Biol. 9:210. doi: 10.1186/1471-2148-9-210
D’Orbigny, A. (1842–1853). “Mollusques,” in Historia Física, Política y Natural de la Isla de Cuba, Vol. 2, ed. R. De La Sagra (Paris: Librería de Arthus Bertrand).
Distel, D. L. (2000). Phylogenetic relationships among Mytilidae (Bivalvia): 18S rRNA data suggest convergence in mytilid body plans. Mol. Phylogenet. Evol. 15, 25–33. doi: 10.1006/mpev.1999.0733
Distel, D. L. (2003). “The Biology of Marine Wood Boring Bivalves And Their Bacterial Endosymbionts,” in Wood Deterioration and Preservation. Wasington: American Chemical Society Publications, 253–271. doi: 10.1021/bk-2003-0845.ch014
Distel, D. L., Amin, M., Burgoyne, A., Linton, E., Mamangkey, G., Morrill, W., et al. (2011). Molecular phylogeny of Pholadoidea Lamarck, 1809 supports a single origin for xylotrophy (wood feeding) and xylotrophic bacterial endosymbiosis in Bivalvia. Mol. Phylogenet Evol. 61, 245–254. doi: 10.1016/j.ympev.2011.05.019
Distel, D. L., Beaudoin, D. J., and Morrill, W. (2002). Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl. Envir. Microbiol. 68, 6292–6299. doi: 10.1128/aem.68.12.6292-6299.2002
Distel, D. L., and Roberts, S. J. (1997). Bacterial endosymbionts in the gills of the deep-sea wood-boring bivalves Xylophaga atlantica and Xylophaga washingtona. Biol. Bull. 192, 253–261. doi: 10.2307/1542719
Dons, C. (1929). Zoologiske Notiser V. Xylophaga dorsalis I Norge. Det Kongelige. norske Vidensk. Selsk. Forh. 65, 196–199.
Dons, C. (1933). Zoologiske notiser XX. Xylophaga praestans, ny for Norges kvartærgeologi. K. norske Vidensk. Selsk. Forh. 5, 191–193.
Emery, W. (2001). Water types and water masses. Encyclop. Ocean Sci. 4, 3179–3187. doi: 10.1006/rwos.2001.0108
Fagervold, S. K., Bessette, S., Romano, C., Martin, D., Plyuscheva, M., Le Bris, N., et al. (2013). Microbial communities associated with the degradation of oak wood in the Blanes submarine canyon and its adjacent open slope (NW Mediterranean). Prog. Oceanogr. 118, 137–143.
Fagervold, S. K., Romano, C., Kalenitchenko, D., Borowski, C., Nunes-Jorge, A., Martin, D., et al. (2014). Microbial communities in sunken wood are structured by wood-boring bivalves and location in a submarine canyon. PLoS One 9:e96248. doi: 10.1371/journal.pone.0096248
Fallén, C. F. (1810). Specim. Entomolog. Novam Diptera Disponendi Methodum Exhibens. Lundae: Berlingianis.
Folmer, O., Black, M., Hoeh, W. W. H., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol 3, 294–299.
Fredj, G., and Laubier, L. (1985). “The deep mediterranean benthos,” in Mediterranean Marine Ecosystems, eds M. Moraitou-Apostolopoulou and V. Kiortsis (Boston, MA: Springer), 109–145. doi: 10.1007/978-1-4899-2248-9_6
Gascard, J. C., and Richez, C. (1985). Water masses and circulation in the Western Alboran sea and in the Straits of Gibraltar. Progr. Oceanogra. 15, 157–216. doi: 10.1016/0079-6611(85)90031-X
Gaudron, S. M., Pradillon, F., Pailleret, M., Duperron, S., Le Bris, N., and Gaill, F. (2010). Colonization of organic substrates deployed in deep-sea reducing habitats by symbiotic species and associated fauna. Mar. Environ. Res. 70, 1–12. doi: 10.1016/j.marenvres.2010.02.002
Gaudron, S. M. T. H., Wang, H., Laming, S. R., and Duperron, S. (2016). Plasticity in reproduction and nutrition in wood-boring bivalves (Xylophaga atlantica) from the Mid-Atlantic Ridge. Mar. Biol 163:213. doi: 10.1007/s00227-016-2988-6
Giribert, G., and Wheeler, W. (2002). On Bivalve phylogeny: a high-level analysis of the Bivalvia (Mollusca) based on combined morphology and DNA sequence data. Invert. Biol. 121, 271–324. doi: 10.1111/j.1744-7410.2002.tb00132.x
Giribet, G., and Distel, D. L. (2003). “Bivalve phylogeny and molecular data,” in Molecular Systematics and Phylogeography of Mollusks, eds C. Lydeard and D. R. Lindberg (Washington, DC: Smithsonian Books), 45–90.
Gray, J. E. (1827). Monograph of the genus Teredo of Linne, with descriptive characters of the species in the British Museum. Phil. Mag. 2, 409–411. doi: 10.1080/14786442708674441
Haderlie, E. C., and Mellor, J. C. (1973). Settlement growth rates and depth preference of the shipworm Bankia setacea in Monterey Bay. Veliger 15, 265–286.
Haga, T., and Kase, T. (2013). Progenetic dwarf males in the deep-sea wood-boring genus Xylophaga (Bivalvia: Pholadoidea). J. Moll. Stud. 79, 90–94. doi: 10.1093/mollus/eys037
Harvey, R. (1996). Deep water Xylophagaidae (Pelecypoda: Pholadacea) from the North Atlantic with descriptions of three new species. J. Conchiol. 35, 473–481.
Hebert, P. D. N., Ratnasingham, S., and Dewaard, J. R. (2003). Barcoding animal life: Cytochrome C Oxidase subunit I divergences among closely related species. Proc. R. Soc. London Ser. B Biol. Sci. 270, S96–S99. doi: 10.1098/rsbl.2003.0025
Hebert, P. D. N., Stoeckle, M. Y., Zemlak, T. S., and Francis, C. M. (2004). Identification of birds through DNA barcodes. PLoS Biol. 2:e312. doi: 10.1371/journal.pbio.0020312
Hsü, K. J., Ryan, W. B. F., and Cita, M. B. (1973). Late Miocene dessication of Mediterranean. Nature 242, 240–244. doi: 10.1038/242240a0
Huelsenbeck, J. P., and Ronquist, F. R. (2001). MrBayes: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
International Commission on Zoological Nomenclature (2018). Opinion 2429 (Case 3717) – Xylophagidae (Mollusca, Bivalvia): emended to Xylophagaidae to remove homonymy with <Xylophagidae Fallén, 1810 (Insecta, Diptera). Bull. Zool. Nomencl. 75, 297–299. doi: 10.21805/bzn.v75.a066
Janssen, R. (1989). Benthos-Mollusken aus dem Tiefenwasser des östliuchen Mittelmeeres, gesammelt während der “METEOR”-Fahrt 5. Sencken. Mar. 20, 265–276.
Janssen, R., and Krylova, E. M. (2014). Deep-sea fauna of European seas: An annotated species check-list of benthic invertebrates living deeper than 2000 m in the seas bordering Europe: bivalvia. Invert. Zool. 11, 43–82. doi: 10.15298/invertzool.11.1.06
Johnson, S. B., Krylova, E. M., Audzijonyte, A., Sahling, H., and Vrijenhoek, R. C. (2017). Phylogeny and origins of chemosynthetic vesicomyid clams. Syst. Biodiv. 15, 346–360. doi: 10.1080/14772000.2016.1252438
Kalenitchenko, D., Fagervold, S. K., Pruski, A. M., Vétion, G., Yücel, M., Le Bris, N., et al. (2015). Temporal and spatial constraints on community assembly during microbial colonization of wood in seawater. ISME J. 9, 2657–2670. doi: 10.1038/ismej.2015.61
Kalenitchenko, D., Le Bris, N., Dadaglio, L., Peru, E., Besserer, A., and Galand, P. E. (2018a). Bacteria alone establish the chemical basis of the wood-fall chemosynthetic ecosystem in the deep-sea. ISME J. 12, 367–379. doi: 10.1038/ismej.2017.163
Kalenitchenko, D., Peru, E., Pereira, L. C., Petetin, C., Galand, P. E., and Le Bris, N. (2018b). The early conversion of deep-sea wood falls into chemosynthetic hotspots revealed by in situ monitoring. Sci. Rep 8:907. doi: 10.1038/s41598-017-17463-2
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Klemm, D., Heublein, B., Fink, H.-P., and Bohn, A. (2005). Cellulose: Fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. 44, 3358–3393. doi: 10.1002/anie.200460587
Knudsen, J. (1961). The bathyal and abissal Xylophaga (Pholadidae. Bivalvia). Galathea Rep. 5, 163–209.
Lane, D. J. (1991). “16S/23S sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley), 115–175.
Leigh, J. W., and Bryant, D. (2015). Popart: Full-feature software for haplotype network construction. Meth. Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Linnaeus, C. (1758). Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, cum characteribus, differentiis, synonymis, locis. Tomus I. Editio Decima, Reformata. Stockholm: Laurentii Salvii. doi: 10.5962/bhl.title.542
MacIntosh, H., De Nys, R., and Whalan, S. (2014). Contrasting life histories in shipworms: Growth, reproductive development and fecundity. J. Exp. Mar. Biol. Ecol. 459, 80–86. doi: 10.1016/j.jembe.2014.05.015
Maddison, W. P., and Maddison, D. R. (2019). Mesquite: A Modular System For Evolutionary Analysis. Version 3.61. Available online at: http://www.mesquiteproject.org (accessed December, 2019).
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Meyer, C. P., and Paulay, G. (2005). DNA Barcoding: Error rates based on comprehensive sampling. PLoS Biol. 3:e422. doi: 10.1371/journal.pbio.0030422
Mienis, H. K. (2003). Woodboring bivalves from shipwrecks in the eastern Mediterranean 2. Xylophaga from the wreck of the submarine “Dakar”. Triton 8, 5–6.
Miller, A. R., Tchernia, P., Charnock, H., and Mcgill, D. A. (1970). Mediterranean Sea Atlas of Temperature, Salinity, Oxygen Profiles And Data From The Cruises of R.V. Atlantis and R.V. Chain With Distribution Of Nutrient Chemical Properties. Woods Hole, MA: Woods Hole Oceanographic Institution.
Monari, S. (2009). Phylogeny and biogeography of pholadid bivalve Barnea (Anchomasa) with considerations on the phylogeny of Pholadoidea. Acta Palaeont. Polonica 54, 315–335. doi: 10.4202/app.2008.0068
O’Connor, R. M., Fung, J. M., Sharp, K. H., Benner, J. S., Mcclung, C., Cushing, S., et al. (2014). Gill bacteria enable a novel digestive strategy in a wood-feeding mollusk. Proc. Nat. Acad. Sci. U.S.A. 111, E5096–E5104. doi: 10.1073/pnas.1413110111
Park, J., and O’Foighil, D. (2000). Sphaeriid and corbiculid clams represent separate heterodont bivalve radiations into freshwater environments. Mol. Phylogenet. Evol. 14, 75–88. doi: 10.1006/mpev.1999.0691
Pérès, J. M. (1985). “History of the Mediterranean biota and the colonization of the depths,” in Key Environments: Western Mediterranean, ed. R. Margalef (Oxford: Pergamon Press), 198–232.
Pop Ristova, P., Bienhold, C., Wenzhofer, F., Rossel, P. E., and Boetius, A. (2017). Temporal and spatial variations of bacterial and faunal communities associated with deep-sea wood falls. PLoS One 12:e169906. doi: 10.1371/journal.pone.0169906
Purchon, R. D. (1941). On the biology and relationships of the lamellibrach Xylophaga dorsalis (Turton). J. Mar. Biol. Ass. U.K. 25, 1–39. doi: 10.1017/S0025315400014259
Rafinesque, C. S. (1815). Analyse de la nature ou Tableau de l’univers et des corps organisés. Palerme: Aux dépens de l’auteur. doi: 10.5962/bhl.title.106607
Rambaut, A. (2006). FigTree v1.3.1. Computer Software and Manual. Available online at: https://tree.bio.ed.ac.uk/software/figtree (accessed July 10, 2017).
Reft, A. J., and Voight, J. R. (2009). Sensory structures on the siphons of wood-boring bivalves (Pholadidae: Xylophagainae: Xylophaga). Nautilus 123, 43–48.
Romano, C., Voight, J. R., Company, J. B., Plyuscheva, M., and Martin, D. (2013). Submarine canyons as the preferred habitat for wood-boring species of Xylophaga (Mollusca. Bivalvia). Prog. Oceanogr. 118, 175–187. doi: 10.1016/j.pocean.2013.07.028
Romano, C., Voight, J. R., Pérez-Portela, R., and Martin, D. (2014). Morphological and genetic diversity of the wood-boring Xylophaga (Mollusca, Bivalvia): new species and records from deep-sea Iberian canyons. PLoS One 9:e102887. doi: 10.1371/journal.pone.0102887
Romey, W. (1991). Recruitment in the deep-sea wood-boring bivalve Xylophaga atlantica Richards. Veliger 34, 14–20.
Romey, W. L., Bullock, R. C., and Dealteris, J. T. (1994). Rapid growth of a deep-sea wood-boring bivalve. Cont. Shelf Res. 14, 1349–1359. doi: 10.1016/0278-4343(94)90052-3
Rosenberg, G., and Gofas, S. (2015). Xylophagidae: MolluscaBase. Available online at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=489105 (accessed July, 2019).
Sabbadin, F., Pesante, G., Elias, L., Besser, K., Li, Y., Steele-King, C., et al. (2018). Uncovering the molecular mechanisms of lignocellulose digestion in shipworms. Biotechnol. Biofuels 11, 59–69. doi: 10.1186/s13068-018-1058-3
Santhakumaran, L. N. (1980). Two new species of Xylophaga from Trondheimsfjorden, western Norway (Mollusca. Pelecypoda). Sarsia 65, 269–272. doi: 10.1080/00364827.1980.10431489
Saraswathy, M., and Nair, N. B. (1971). XIV.—Observations on the structure of the shipworms, Nausitora hedleyi, Teredo furcifera and Teredora princesae (Bivalvia: Teredinidae). Trans. R. Soc. Edinburgh 68, 507–566. doi: 10.1017/S0080456800014861
Sharma, P. P., Zardus, J. D., Boyle, E. E., González, V. L., Jennings, R. M., Mcintyre, E., et al. (2013). Into the deep: a phylogenetic approach to the bivalve subclass Protobranchia. Mol. Phylogenet. Evol. 69, 188–204. doi: 10.1016/j.ympev.2013.05.018
Smith, C. R., De Leo, F. C., Bernardino, A. F., Sweetman, A. K., and Martínez Arbizu, P. (2008). Abyssal food limitation, ecosystem structure and climate change. Trends Ecol. Evol. 23, 518–528. doi: 10.1016/j.tree.2008.05.002
Smith, E. A. (1904). On a collection of marine shells from Port Alfred. Cape Colony. J. Malacol. 11, 21–44.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stockton, W. L., and DeLaca, T. E. (1982). Food falls in the deep sea: occurrence, quality, and significance. Deep-Sea Res. A Oceanogr. Res. Pap 29, 157–169. doi: 10.1016/0198-0149(82)90106-6
Swofford, D. L. (2003). PAUP∗. Phylogenetic analysis using parsimony (∗ and other methods). Version 4.0. Sunderland: Sinauer Associates.
Taki, I., and Habe, T. (1950). Xylophaginidae in Japan. Illust. Cat. Jap. Shells. 7, 45–47. doi: 10.6013/jbrewsocjapan1915.45.3_47
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Turner, R. D. (1955). The family Pholadidae in the western Atlantic and the eastern Pacific. Part II – Martesiinae Jouannetiinae Xylopagainae. Johnsonia 3, 65–160.
Turner, R. D. (1972). Xyloredo, a new teredinid-like abyssal wood-borer (Mollusca, Pholadidae, Xylophagainae). Breviora 397, 1–19.
Turner, R. D. (1973). Wood-boring bivalves, opportunistic species in the deep sea. Science 180, 1377–1379. doi: 10.1126/science.180.4093.1377
Turner, R. D. (2002). On the subfamily Xylophagainae (Family Pholadidae. Bivalvia, Mollusca). Bull. Museum Comp. Zool. 157, 223–308.
Turner, R. D., and Culliney, J. (1971). Some anatomical and life history studies of wood boring bivalve systematics. Rep. Am. Malacol. Un. Pacific Div. 1970, 65–66.
Turton, W. (1819). A Conchological Dictionary of the British Islands. London: John Booth. doi: 10.5962/bhl.title.156842
Turton, W. (1822). Conchylia insularum britannicarum. London: Nattali and Combe. doi: 10.5962/bhl.title.10443
Tyler, P. A., Young, C. M., and Dove, F. (2007). Settlement, growth and reproduction in the deep-sea wood-boring bivalve mollusc Xylophaga depalmai. Mar. Ecol. Progr. Ser. 343, 151–159. doi: 10.3354/meps06832
Tyler, P. A., and Young, G. M. (1999). Reproduction and dispersal at vents and cold seeps. J. Mar. Biol. Ass. U.K. 79, 193–208. doi: 10.1017/S0025315499000235
Van Dover, C. L., German, C. R., Speer, K. G., Parson, L. M., and Vrijenhoek, R. C. (2002). Evolution and biogeography of deep-sea vent and seep invertebrates. Science 295:1253. doi: 10.1126/science.1067361
Voight, J. R. (2007). Experimental deep-sea deployments reveal diverse Northeast Pacific wood-boring bivalves of Xylophagainae (Myoida : Pholadidae). J. Moll. Stud. 73, 377–391. doi: 10.1093/mollus/eym034
Voight, J. R. (2008). Deep-sea wood-boring bivalves of Xylophaga (Myoida: Pholadidae) on the Continental Shelf: a new species described. J. Mar. Biol. Ass. U.K. 88, 1459–1464. doi: 10.1017/S0025315408002117
Voight, J. R. (2009). Diversity and reproduction of near-shore vs offshore wood-boring bivalves (Pholadidae: Xylophagainae) of the deep eastern Pacific ocean, with three new species. J. Moll. Stud. 75, 167–174. doi: 10.1093/mollus/eyp012
Voight, J. R. (2015). Xylotrophic bivalves: aspects of their biology and the impacts of humans. J. Moll. Stud. 81, 175–186. doi: 10.1093/mollus/eyv008
Voight, J. R. (2016). New insights on Xylopholas (Mollusca: Xylophagaidae): diversity, growth and reproduction. Am. Malacol. Bull. 34, 138–146. doi: 10.4003/006.034.0210
Voight, J. R., Jacob, C. C., and Raymond, W. L. (2020). Stable isotopic evidence of mixotrophy in xylophagaids, deep-sea wood-boring bivalves. Front. Mar. Sci 7:50. doi: 10.3389/fmars.2020.00050
Voight, J. R., Marshall, B. A., Judge, J., Halanych, K. M., Li, Y., Bernardino, A. F., et al. (2019). Life in wood: preliminary phylogeny of deep-sea wood-boring bivalves (Xylophagaidae), with descriptions of three new genera and one new species. J. Moll. Stud. 85, 232–243. doi: 10.1093/mollus/eyz003
Voight, J. R., and Segonzac, M. (2012). At the bottom of the deep blue sea: a new wood-boring bivalve (Mollusca. Pholadidae, Xylophaga) from the Cape Verde byssal Plain (subtropical Atlantic). Zoosystema 34, 171–180. doi: 10.5252/z2012n1a8
Vrijenhoek, R. C., Johnson, S. B., and Rouse, G. W. (2009). A remarkable diversity of bone-eating worms (Osedax; Siboglinidae; Annelida). BMC Biol. 7:74. doi: 10.1186/1741-7007-7-74
Waterbury, J. B., Calloway, C. B., and Turner, R. D. (1983). A cellulolytic nitrogen-fixing bacterium cultured from the gland of Deshayes in shipworms (Bivalvia: Teredinidae). Science 221, 1401–1403. doi: 10.1126/science.221.4618.1401
Weigelt, R., Lippert, H., Borges, L. M. S., Appelqvist, C., Karsten, U., and Bastrop, R. (2016). First time DNA barcoding of the common shipworm Teredo navalis Linnaeus, 1758 (Mollusca: Bivalvia: Teredinidae): molecular-taxonomic investigation and identification of a widespread wood-borer. J. Exp. Mar. Biol. Ecol. 475, 154–162. doi: 10.1016/j.jembe.2015.11.008
West, A. J., Lin, C. W., Lin, T. C., Hilton, R. G., Liu, S. H., Chang, C. T., et al. (2011). Mobilization and transport of coarse woody debris to the oceans triggered by an extreme tropical storm. Limnol. Oceanogr. 56, 77–85. doi: 10.4319/lo.2011.56.1.0077
Wolff, T. (1979). Macrofaunal utilization of plant remains in the deep-sea. Sarsia 64, 117–136. doi: 10.1080/00364827.1979.10411373
Yücel, M., Galand, E., Fagervold, S. A. K., Contreira-Pereira, L., and Le Bris, N. (2013). Sulfide production and consumption in degrading wood in the marine environment. Chemosphere 90, 403–409. doi: 10.1016/j.chemosphere.2012.07.036
Zenetos, A., Vardala-Theodoro, E., and Alexandrakis, C. (2005). Update of the marine Bivalvia Mollusca checklist in Greek waters. J. Mar. Biol. Ass. U.K. 85, 993–998. doi: 10.1017/S0025315405012014
Keywords: genetic connectivity, wood falls, Mediterranean, deep sea, phylogeography
Citation: Romano C, Nunes-Jorge A, Le Bris N, Rouse GW, Martin D and Borowski C (2020) Wooden Stepping Stones: Diversity and Biogeography of Deep-Sea Wood Boring Xylophagaidae (Mollusca: Bivalvia) in the North-East Atlantic Ocean, With the Description of a New Genus. Front. Mar. Sci. 7:579959. doi: 10.3389/fmars.2020.579959
Received: 03 July 2020; Accepted: 29 September 2020;
Published: 20 November 2020.
Edited by:
Marcos Rubal, University of Porto, PortugalReviewed by:
Gonzalo Giribet, Harvard University, United StatesAnna Marie Holmes, National Museum Wales, United Kingdom
Copyright © 2020 Romano, Nunes-Jorge, Le Bris, Rouse, Martin and Borowski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiara Romano, cromano@ceab.csic.es; Christian Borowski, cborowsk@mpi-bremen.de
†ORCID: Chiara Romano, orcid.org/0000-0001-5078-0082; Nadine Le Bris, orcid.org/0000-0002-0142-4847; Greg W. Rouse, orcid.org/0000-0001-9036-9263; Daniel Martin, orcid.org/0000-0001-6350-7384; Christian Borowski, orcid.org/0000-0001-7921-3022
 Chiara Romano
Chiara Romano Amandine Nunes-Jorge
Amandine Nunes-Jorge Nadine Le Bris
Nadine Le Bris Greg W. Rouse
Greg W. Rouse Daniel Martin
Daniel Martin Christian Borowski
Christian Borowski