Abstract
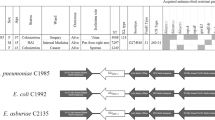
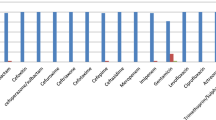
In the last years, an increasing number of untreatable infections caused by drug-resistant microbes have impacted the health care system. Worldwide, infections caused by carbapenem-resistant (CR) Gram-negative bacilli have dramatically increased. Among the CR-Gram-negative bacilli, those producing carbapenemases, such as NDM-1, are the main concern. Different Enterobacterales harboring NDM-1 have been reported lately. Providencia stuartii, a member of the Morganellaceae family, is ubiquitous in the environment, but is also known to cause nosocomial infections. Here we describe the genomic analysis of two NDM-1- producing P. stuartii strains recovered from the same patient as well as other carbapenem resistant strains recovered from the same hospital. As a result of the genomic analysis thirteen resistance genes, including three to β-lactams (blaOXA-1, blaTEM-1, blaNDM-1), four to aminoglycosides (aphA6, aac(3)-IId, aac(2′)-Ia, aac(6′)-Ib-cr5), one to sulfonamides (sul1), two to chloramphenicol (catB3, catA3), one to rifampicin, one to bleomycin (ble), and one to tetracycline (tet(B)) were found. Moreover, a variety of mobile genetic elements, such as insertion sequences, plasmids and phage- related sequences, were found within P. stuartii genomes. The spread of carbapenem-resistant isolates remains a significant clinical and public health concern. Therefore, we considered that the detection of CR isolates is an essential step in addressing this problem.


Similar content being viewed by others
References
Durante-Mangoni E, Andini R, Zampino R (2019) Management of carbapenem-resistant Enterobacteriaceae infections. Clin Microbiol Infect 25:943–950
Lutgring JD (2019) Carbapenem-resistant Enterobacteriaceae: An emerging bacterial threat. Semin Diagn Pathol 36:182–186
Yoo JH (2018) The Infinity War: How to Cope with Carbapenem-resistant Enterobacteriaceae. J Korean Med Sci 33:e255
Nordmann P, Naas T, Poirel L (2011) Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798
Martinez-Martinez L (2019) Carbapenemases: The never-ending story. Enferm Infecc Microbiol Clin 37:73–75
Abdallah M, Balshi A (2018) First literature review of carbapenem-resistant Providencia. New Microbes New Infect 25:16–23
Wenzel RP, Hunting KJ, Osterman CA, Sande MA (1976) Providencia stuartii, a hospital pathogen: potential factors for its emergence and transmission. Am J Epidemiol 104:170–180
Krake PR, Tandon N (2004) Infective endocarditis due to Providenca stuartii. South Med J 97:1022–1023
Sipahi OR, Bardak-Ozcem S, Ozgiray E, Aydemir S, Yurtseven T, Yamazhan T, Tasbakan M, Ulusoy S (2010) Meningitis due to Providencia stuartii. J Clin Microbiol 48:4667–4668
Unverdi S, Akay H, Ceri M, Inal S, Altay M, Demiroz AP, Duranay M (2011) Peritonitis due to Providencia stuartii. Perit Dial Int 31:216–217
Woods TD, Watanakunakorn C (1996) Bacteremia due to Providencia stuartii: review of 49 episodes. South Med J 89:221–224
Manageiro V, Sampaio DA, Pereira P, Rodrigues P, Vieira L, Palos C, Canica M. 2015. Draft Genome Sequence of the First NDM-1-Producing Providencia stuartii Strain Isolated in Portugal. Genome Announc 3.
McGann P, Hang J, Clifford RJ, Yang Y, Kwak YI, Kuschner RA, Lesho EP, Waterman PE (2012) Complete sequence of a novel 178-kilobase plasmid carrying bla(NDM-1) in a Providencia stuartii strain isolated in Afghanistan. Antimicrob Agents Chemother 56:1673–1679
Molnar S, Flonta MMM, Almas A, Buzea M, Licker M, Rus M, Foldes A, Szekely E (2019) Dissemination of NDM-1 carbapenemase-producer Providencia stuartii strains in Romanian hospitals: a multicentre study. J Hosp Infect 103:165–169
Aires CAM, Almeida ACS, Vilela MA, Morais-Junior MA, Morais MMC (2016) Selection of KPC-2-producing Providencia stuartii during treatment for septicemia. Diagn Microbiol Infect Dis 84:95–96
Douka E, Perivolioti E, Kraniotaki E, Fountoulis K, Economidou F, Tsakris A, Skoutelis A, Routsi C (2015) Emergence of a pandrug-resistant VIM-1-producing Providencia stuartii clonal strain causing an outbreak in a Greek intensive care unit. Int J Antimicrob Agents 45:533–536
Miriagou V, Tzouvelekis LS, Flevari K, Tsakiri M, Douzinas EE (2007) Providencia stuartii with VIM-1 metallo-beta-lactamase. J Antimicrob Chemother 60:183–184
Oikonomou O, Liakopoulos A, Phee LM, Betts J, Mevius D, Wareham DW (2016) Providencia stuartii Isolates from Greece: Co-Carriage of Cephalosporin (blaSHV-5, blaVEB-1), Carbapenem (blaVIM-1), and Aminoglycoside (rmtB) Resistance Determinants by a Multidrug-Resistant Outbreak Clone. Microb Drug Resist 22:379–386
Tavares CP, Pereira PS, Marques Ede A, Faria C Jr, de Souza MP, de Almeida R, Alves Cde F, Asensi MD, Carvalho-Assef AP (2015) Molecular epidemiology of KPC-2-producing Enterobacteriaceae (non-Klebsiella pneumoniae) isolated from Brazil. Diagn Microbiol Infect Dis 82:326–330
Institute CaLS. 2020. Performance Standards for Antimicrobial Susceptibility Testing; M.100., twenty-Nine Informational Supplement.
Pires J, Novais A, Peixe L (2013) Blue-carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol 51:4281–4283
Pantel A, Souzy D, Sotto A, Lavigne JP (2015) Evaluation of Two Phenotypic Screening Tests for Carbapenemase-Producing Enterobacteriaceae. J Clin Microbiol 53:3359–3362
Montana S, Palombarani S, Carulla M, Kunst A, Rodriguez CH, Nastro M, Vay C, Ramirez MS, Almuzara M (2018) First case of bacteraemia due to Acinetobacter schindleri harbouring blaNDM-1 in an immunocompromised patient. New Microbes New Infect 21:28–30
Montana S, Schramm ST, Traglia GM, Chiem K, Parmeciano Di Noto G, Almuzara M, Barberis C, Vay C, Quiroga C, Tolmasky ME, Iriarte A, Ramirez MS (2016) The Genetic Analysis of an Acinetobacter johnsonii Clinical Strain Evidenced the Presence of Horizontal Genetic Transfer. PLoS ONE 11:e0161528
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain J-M (2014) ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob Agents Chemother 58:212–220
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FS, Wright GD, McArthur AG (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573
Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M (2006) ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36
Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS (2011) PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Martino F, Tijet N, Melano R, Petroni A, Heinz E, De Belder D, Faccone D, Rapoport M, Biondi E, Rodrigo V, Vazquez M, Pasteran F, Thomson NR, Corso A, Gomez SA (2019) Isolation of five Enterobacteriaceae species harbouring blaNDM-1 and mcr-1 plasmids from a single paediatric patient. PLoS ONE 14:e0221960
Aires-de-Sousa M, Ortiz de la Rosa JM, Goncalves ML, Costa A, Nordmann P, Poirel L (2020) Occurrence of NDM-1-producing Morganella morganii and Proteus mirabilis in a single patient in Portugal: probable in vivo transfer by conjugation. J Antimicrob Chemother 75:903–906
Abdallah M, Alhababi R, Alqudah N, Aldyyat B, Alharthy A (2018) First report of carbapenem-resistant Providencia stuartii in Saudi Arabia. New Microbes New Infect 26:107–109
Mao YC, Chang CL, Huang YC, Su LH, Lee CT (2018) Laboratory investigation of a suspected outbreak caused by Providencia stuartii with intermediate resistance to imipenem at a long-term care facility. J Microbiol Immunol Infect 51:214–219
Mnif B, Ktari S, Chaari A, Medhioub F, Rhimi F, Bouaziz M, Hammami A (2013) Nosocomial dissemination of Providencia stuartii isolates carrying blaOXA-48, blaPER-1, blaCMY-4 and qnrA6 in a Tunisian hospital. J Antimicrob Chemother 68:329–332
Saida NB, Thabet L, Messadi A, Bouselmi K, Turki A, Boukadida J (2008) Clonality of Providencia stuartii isolates involved in outbreak that occurred in a burn unit. Burns 34:829–834
Zavascki AP, Carvalhaes CG, da Silva GL, Tavares Soares SP, de Alcantara LR, Elias LS, Sandri AM, Gales AC (2012) Outbreak of carbapenem-resistant Providencia stuartii in an intensive care unit. Infect Control Hosp Epidemiol 33:627–630
Clifford RJ, Hang J, Riley MC, Onmus-Leone F, Kuschner RA, Lesho EP, Waterman PE (2012) Complete genome sequence of Providencia stuartii clinical isolate MRSN 2154. J Bacteriol 194:3736–3737
Liakopoulos A, Oikonomou O, Wareham DW. 2017. Draft Genome Sequence of Providencia stuartii PS71, a Multidrug-Resistant Strain Associated with Nosocomial Infections in Greece. Genome Announc 5.
Pillonetto M, Arend L, Vespero EC, Pelisson M, Chagas TP, Carvalho-Assef AP, Asensi MD (2014) First report of NDM-1-producing Acinetobacter baumannii sequence type 25 in Brazil. Antimicrob Agents Chemother 58:7592–7594
Tumbarello M, Citton R, Spanu T, Sanguinetti M, Romano L, Fadda G, Cauda R (2004) ESBL-producing multidrug-resistant Providencia stuartii infections in a university hospital. J Antimicrob Chemother 53:277–282
Politi L, Gartzonika K, Spanakis N, Zarkotou O, Poulou A, Skoura L, Vrioni G, Tsakris A (2019) Emergence of NDM-1-producing Klebsiella pneumoniae in Greece: evidence of a widespread clonal outbreak. J Antimicrob Chemother 74:2197–2202
Kanzari L, Ferjani S, Saidani M, Hamzaoui Z, Jendoubi A, Harbaoui S, Ferjani A, Rehaiem A, Boutiba Ben Boubaker I, Slim A (2018) First report of extensively-drug-resistant Proteus mirabilis isolate carrying plasmid-mediated blaNDM-1 in a Tunisian intensive care unit. Int J Antimicrob Agents 52:906–909
Shin S, Jeong SH, Lee H, Hong JS, Park MJ, Song W (2018) Emergence of multidrug-resistant Providencia rettgeri isolates co-producing NDM-1 carbapenemase and PER-1 extended-spectrum beta-lactamase causing a first outbreak in Korea. Ann Clin Microbiol Antimicrob 17:20
Marquez-Ortiz RA, Haggerty L, Olarte N, Duarte C, Garza-Ramos U, Silva-Sanchez J, Castro BE, Sim EM, Beltran M, Moncada MV, Valderrama A, Castellanos JE, Charles IG, Vanegas N, Escobar-Perez J, Petty NK (2017) Genomic Epidemiology of NDM-1-Encoding Plasmids in Latin American Clinical Isolates Reveals Insights into the Evolution of Multidrug Resistance. Genome Biol Evol 9:1725–1741
Plante I, Centron D, Roy PH (2003) An integron cassette encoding erythromycin esterase, ere(A), from Providencia stuartii. J Antimicrob Chemother 51:787–790
Barlow RS, Pemberton JM, Desmarchelier PM, Gobius KS (2004) Isolation and characterization of integron-containing bacteria without antibiotic selection. Antimicrob Agents Chemother 48:838–842
Barlow RS, Gobius KS (2006) Diverse class 2 integrons in bacteria from beef cattle sources. J Antimicrob Chemother 58:1133–1138
Acknowledgements
Part of the authors’ work was supported by NIH SC3GM125556 to MSR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.S.F. was supported by grant MHIRT 2T37MD001368 from the National Institute on Minority Health and Health Disparities, National Institute of Health. SM is recipient of a postdoctoral fellowship from CONICET. C.Q and G.M.T. are members of the CIC from CONICET.
Author information
Authors and Affiliations
Contributions
(1) Conception and design of the study: AH, SM, AM, JSF, GMT, CQ, MA, MSR; (2) Acquisition of data: AH, SM, AM, JSF, GMT, AF, EC, CC, MA, MSR; (3) Analysis of data: AH, SM, GMT, CQ, MSR; (4) Drafting and revision of manuscript: AH, SM, GMT, CQ, MA, MSR.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that they have no conflict of interest.
Ethical Standards
All procedures performed in this study were in accordance with the ethical standards of the Hospital Eva Peron, Buenos Aires, Argentina, the 1964 Helsinki Declaration, its later amendments and the National Law on the protection of personal data No 25.326.
Informed Consent
Informed consent was obtained from the patient involved in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hoard, A., Montaña, S., Moriano, A. et al. Genomic Analysis of two NDM-1 Providencia stuartii Strains Recovered from a Single Patient. Curr Microbiol 77, 4029–4036 (2020). https://doi.org/10.1007/s00284-020-02242-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02242-6