Abstract
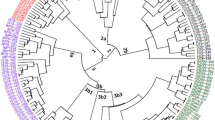
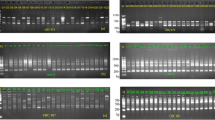
Oil palm (Elaeis guineensis Jacq.) is the key vegetable oil yielded crop and widely distributed in the southern region of China. The present study aimed at analyzing the genetic diversity and population structure of 223 oil palm accessions collected from four provinces of China by using the sequence-related amplified polymorphism (SRAP) markers. A set of 33 SRAP molecular markers were employed to analyze genetic diversity as well as population structure across 223 oil palm accessions. Out of 514 amplified bands, 487 (94.75%) bands were found to be polymorphic. The PIC value (polymorphic information content) of detected bands was from 0.38 to 0.51, with 0.46 of average. The STRUCTURE analysis categorized the 223 oil palm accessions into three subpopulations. The UPGMA based clustering classified them into three major clusters. The correlation between the population subdivisions and the genetic relationships among oil palm accessions was revealed by UPGMA and Bayesian STRUCTURE analyses. The principal coordinate analysis also confirmed a similar grouping of accessions as revealed by the UPGMA dendrogram and STRUCTURE analysis. AMOVA analysis also revealed the variance of 24% among subpopulations and 76% within subpopulations. The present investigation provided valuable information on population structure and genetic diversity of oil palm populations in China for molecular breeding research in oil palm.




Similar content being viewed by others
Data Availability
The SRAP (Sequence related amplified polymorphism) primers and SRAP data are available in the article and oil palm accessions used in this study are given in the supplementary table.
Abbreviations
- SRAP:
-
Sequence-Related Amplified Polymorphism
- DNA:
-
deoxyribonucleic acid
References
Agbagwa I, Patil P, Datta S, Nadarajan N (2014) Comparative efficiency of SRAP, SSR and AFLP-RGA markers in resolving genetic diversity in pigeon pea (Cajanus sp.). Ind J Biotech 13:486–495
Agyenim-Boateng KG, Lu JN, Shi YZ, Zhang D, Yin XG (2019) SRAP analysis of the genetic diversity of wild castor (Ricinus communis L.) in South China. PLoS One 14(7):e0219667
Bai B, Wang L, Zhang YJ, Lee M, Rahmadsyah R, Alfiko Y, Ye BQ, Purwantomo S, Suwanto A, Chua NH, Yue GH (2018) Developing genome-wide SNPs and constructing an ultrahigh-density linkage map in oil palm. Sci Rep 8:691–695
Bhatt J, Kumar S, Patel S, Solanki R (2017) Sequence-related amplified polymorphism (SRAP) markers based genetic diversity analysis of cumin genotypes. Ann Agrar Sci 15:434–438
Ding C, Lin WF (2010) ISSR analysis on genetic diversity of new oil palm germplasm. J Anhui Agric Sci 38(4):2202–2205
Ding C, Lang NJ, Lin WF (2012) Study on genetic diversity of 12 oil palm varieties introduced newly. J Chin Cereals Oils Assoc 27(5):60–69
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Frankel OH, Brown AHD (1984) Plant genetic resources today: a critical appraisal. In: Holden JHW, Williams JT (eds) Crop genetic resources: conservation and evaluation. Academic, London, pp 249–257
Ganesh SK, Thangavelu S (1995) Genetic divergence in sesame (Sesamum indicum L.). Madras Agric J 82:263–265
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives. Genet Res Inter 43:1487
Hanna BB, Malgorzate T, Leszek B, Andrzej K, Monika RT (2014) Genome-wide characterization of genetic diversity and population structure in Secale. BMC Plant Biol 14:184–198
Lei XT, Cao HX (2013) Oil palm. China Agricult Press, Beijing
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor App Genet 103:455–461
Li BJ, Wang JY, Liu ZJ, Zhuang XY, Huang JX (2018a) Genetic diversity and ex-situ conservation of loropetalum subcordatum, an endangered species endemic to China. BMC Genet 19:12
Li SF, Lang XD, Liu WD, Ou GL, Xu H, Su JR (2018) The relationship between species richness and aboveground biomass in a primary Pinus kesiya forest of Yunnan, southwestern China. PLoS One 13:e0191140. https://doi.org/10.1371/journal.pone.0191140
Liu C, White M, Newell G, Pearson R (2013) Selecting thresholds for the prediction of species occurrence with presence-only data. J Biogeogra 40(4):778–789
Liu L, Chen W, Zheng X, Li J, Yan DT, Liu L, Liu X, Wang YL (2016) Genetic diversity of Ulmus lamellosa by morphological traits and sequence-related amplified polymorphism (SRAP) markers. Biochem Syst Ecol 66:272–280
Ma Y, Qu SQ, Xu XR, Liang TT, Zhang DK (2015) Genetic diversity analysis of Cotoneaster schantungensis Klotz. Using SRAP marker. Ameri J Plant Sci 6:2860–2866
Mahendrasinh CT, Ranbir SF, Sushil K, Amar AS (2019) Sequence-related amplified polymorphism (SRAP) analysis of teak (Tectona grandis L.) germplasm. Eco Genet Genom 12:100041
Maizura I, Rajanaidu N, Zakri AH (2006) Assessment of genetic diversity in oil palm (Elaeis guineensis Jacq.) using restriction fragment length polymorphism (RFLP). Genet Resour Crop Evol 53:187–195
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Moretzsohn MC, Nunes CDM, Ferreira ME (2000) RAPD linkage mapping of the shell thickness locus in oil palm (Elaeis guineensis). Theor Appl Genet 100(1):63–70
Ong PW, Maizura I, Abdullah NA, Rafii MY, Ooi LC, Low ET, Singh R (2015) Development of SNP markers and their application for genetic diversity analysis in the oil palm (Elaeis guineensis). Genet Mol Res 14(4):12205–12216
Peakall ROD, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Premkrishnan BV, Arunachalam V (2012) In silico RAPD priming sites in expressed sequences and iSCAR markers for oil palm. Comp Funct Genomics 9:13709
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rance K, Mayes S, Price Z, Jack PL, Corley RHV (2001) Quantitative trait loci for yield components in oil palm (Elaeis guineensis Jacq.). Theor App Genet 103:1302–1310
Rohlf FJ (2000) NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.1. Exeter Software, Setauket
Shaye NA, Migdadi H, Charbaji A, Alsayegh S, Daoud S, Al-Anazi W, Alghamdi S (2018) Genetic variation among saudi tomato (solanum lycopersicum L.) landraces studied using sds-page and srap markers. Saudi J Biol Sci 4:1–9
Shevade VS, Loboda TV (2019) Oil palm plantations in Peninsular Malaysia: Determinants and constraints on expansion. PLoS One 14(2):e0210628
Singh R, Nagappan J, Tan SG, Panandam JM, Cheah SC (2007) Development of simple sequence repeat (SSR) markers for oil palm and their application in genetic mapping and fingerprinting of tissue culture clones. Asia Pacific J Mol Biol biotech 15(3):121–131
Stewart C, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–750
Tamaki I, Okada M (2014) Genetic admixing of two evergreen oaks, Quercus acuta and Q. sessilifolia (subgenus Cyclobalanopsis), is the result of interspecific introgressive hybridization. Tree Genet Genomes 10:989–999
Utomo C, Werner S, Niepold F, Deising HB (2005) Identification of Ganoderma, the causal agent of basal stem rot disease in oil palm using a molecular method. Mycopathol 159(1):159–170
Wang D, Shen B, Gong H (2019) Genetic diversity of Simao pine in China revealed by SRAP markers. PeerJ 7:e6529
Xia W, Luo TT, Zhang W, Mason AS, Huang DY, Huang XL, Tang WQ, Zhang CY, Xiao Y (2019) Development of high-density SNP markers and their application in evaluating genetic diversity and population structure in Elaeis guineensis. Front Plant Sci 10:130–141
Xiao Y, Zhou LX, Xia W, Mason AS, Yang YD, Ma ZL, Peng ME (2014) Exploiting transcriptome data for the development and characterization of gene-based SSR markers related to cold tolerance in oil palm (Elaeis guineensis). BMC Plant Biol 14:384–397
Yarra R, Jin L, Zhao Z, Cao H (2019) Progress in tissue culture and genetic transformation of oil palm: an overview. Int J Mol Sci 20(21):5353
Youssef M, James AC, Rivera-Madrid R et al (2011) Musa genetic diversity revealed by SRAP and AFLP. Mol Biotechnol 47:189–199
Yuan XY, Sun YW, Bai XR, Dang M, Feng XJ, Zulfiqar S, Zhao P (2018) Population structure, genetic diversity, and gene introgression of two closely related walnuts (Juglans regia and J. sigillata) in southwestern China revealed by EST-SSR markers. Forests 9:646
Zagorcheva T, Stanev S, Rusanov K, Atanassov I (2020) SRAP markers for genetic diversity assessment of lavender (Lavandula angustifolia mill.) varieties and breeding lines. Biotechnol Biotechnol Equip 34:(1):303–308
Zheng Y, Xu S, Liu J, Zhao Y, Liu J (2017) Genetic diversity and population structure of Chinese natural bermudagrass [Cynodon dactylon (L.) Pers.] germplasm based on SRAP markers. PLoS One 12(5):e0177508
Zhou LX, Xiao Y, Xia W, Yang YD (2015) Analysis of genetic diversity and population structure of oil palm (Elaeis guineensis) from China and Malaysia based on species-specific simple sequence repeat markers. Genet Mol Res 14(4):16247–16254
Zhu SS, Zhu ZY, Wang HR, Wang L, Cheng LZ, Yuan YJ, Zheng YS, Li DD (2018) Characterization and functional analysis of a plastidial FAD6 gene and its promoter in the mesocarp of oil palm (Elaeis guineensis). Sci Hortic 239:163–170
Acknowledgements
This work was supported by the Hainan Provincial Natural Science Foundation of China (319QN324), the Central public-interest scientific institution basal research fund for the Chinese Academy of Tropical Agricultural Sciences (No.1630152019001 and No.1630152017008), the fund for “The fund for species and varieties conservation of sector project of the Ministry of Agriculture and Rural Affairs”. (18200039).
Author information
Authors and Affiliations
Contributions
LZ and HC conceived and designed the experiments. LZ and RY performed the experiments; LZ and RY validated the data, LZ and RY performed the formal analysis; LZ, RY and HC participated in original draft preparation; HC supervised the research.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by: Sithichoke Tangphatsornruang
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhou, L., Yarra, R., Cao, H. et al. Sequence-Related Amplified Polymorphism (SRAP) Markers Based Genetic Diversity and Population Structure Analysis of Oil Palm (Elaeis guineensis Jacq.). Tropical Plant Biol. 14, 63–71 (2021). https://doi.org/10.1007/s12042-020-09273-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-020-09273-0