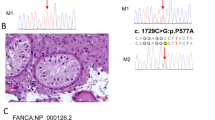
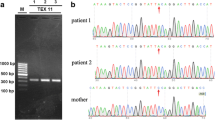
Abstract
Non-obstructive azoospermia (NOA), the lack of spermatozoa in semen due to impaired spermatogenesis affects nearly 1% of men. In about half of cases, an underlying cause for NOA cannot be identified. This study aimed to identify novel variants associated with idiopathic NOA. We identified a nonconsanguineous family in which multiple sons displayed the NOA phenotype. We performed whole-exome sequencing in three affected brothers with NOA, their two unaffected brothers and their father, and identified compound heterozygous frameshift variants (one novel and one extremely rare) in Telomere Repeat Binding Bouquet Formation Protein 2 (TERB2) that segregated perfectly with NOA. TERB2 interacts with TERB1 and Membrane Anchored Junction Protein (MAJIN) to form the tripartite meiotic telomere complex (MTC), which has been shown in mouse models to be necessary for the completion of meiosis and both male and female fertility. Given our novel findings of TERB2 variants in NOA men, along with the integral role of the three MTC proteins in spermatogenesis, we subsequently explored exome sequence data from 1495 NOA men to investigate the role of MTC gene variants in spermatogenic impairment. Remarkably, we identified two NOA patients with likely damaging rare homozygous stop and missense variants in TERB1 and one NOA patient with a rare homozygous missense variant in MAJIN. Available testis histology data from three of the NOA patients indicate germ cell maturation arrest, consistent with mouse phenotypes. These findings suggest that variants in MTC genes may be an important cause of NOA in both consanguineous and outbred populations.



Similar content being viewed by others
Change history
06 February 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00439-020-02244-1
References
Alhathal N, Maddirevula S, Coskun S et al (2020) A genomics approach to male infertility. Genet Med. https://doi.org/10.1038/s41436-020-0916-0
Boivin J, Bunting L, Collins JA, Nygren KG (2007) International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum Reprod 22:1506–1512. https://doi.org/10.1093/humrep/dem046
Djureinovic D, Fagerberg L, Hallström B et al (2014) The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol Hum Reprod 20:476–488. https://doi.org/10.1093/molehr/gau018
Dunce JM, Milburn AE, Gurusaran M et al (2018) Structural basis of meiotic telomere attachment to the nuclear envelope by MAJIN-TERB2-TERB1. Nat Commun 9:1–18. https://doi.org/10.1038/s41467-018-07794-7
Ferlin A, Raicu F, Gatta V et al (2007) Male infertility: role of genetic background. Reprod Biomed Online 14:734–745. https://doi.org/10.1016/S1472-6483(10)60677-3
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907
Glazer CH, Bonde JP, Giwercman A et al (2017) Risk of diabetes according to male factor infertility: a register-based cohort study. Hum Reprod 32:1474–1481. https://doi.org/10.1093/humrep/dex097
Houston BJ, Conrad DF, O’Bryan MK (2020) A framework for high-resolution phenotyping of candidate male infertility mutants: from human to mouse. Hum Genet. https://doi.org/10.1007/s00439-020-02159-x
Hu Z, Yau C, Ahmed AA (2017) A pan-cancer genome-wide analysis reveals tumour dependencies by induction of nonsense-mediated decay. Nat Commun 8:1–9. https://doi.org/10.1038/ncomms15943
Karczewski KJ, Francioli LC, Tiao G et al (2019) Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019:531210. https://doi.org/10.1101/531210
Kasak L, Laan M (2020) Monogenic causes of non-obstructive azoospermia: challenges, established knowledge, limitations and perspectives. Hum Genet. https://doi.org/10.1007/s00439-020-02112-y
Kasak L, Punab M, Nagirnaja L et al (2018) Bi-allelic recessive loss-of-function variants in FANCM cause non-obstructive azoospermia. Am J Hum Genet 103:200–212. https://doi.org/10.1016/j.ajhg.2018.07.005
Kliesch S (2014) Diagnosis of male infertility: diagnostic work-up of the infertile man. Eur Urol Suppl 13:73–82. https://doi.org/10.1016/j.eursup.2014.08.002
Krausz C, Riera-Escamilla A (2018) Genetics of male infertility. Nat Rev Urol 15:369–384. https://doi.org/10.1038/s41585-018-0003-3
Krausz C, Hoefsloot L, Simoni M, Tüttelmann F (2014) EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology 2:5–19
Krausz C, Riera-Escamilla A, Moreno-Mendoza D et al (2020) Genetic dissection of spermatogenic arrest through exome analysis: clinical implications for the management of azoospermic men. Genet Med. https://doi.org/10.1038/s41436-020-0907-1
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. https://doi.org/10.1093/bioinformatics/btp698
Martinez G, Kherraf Z-E, Zouari R et al (2018) Whole-exome sequencing identifies mutations in FSIP2 as a recurrent cause of multiple morphological abnormalities of the sperm flagella. Hum Reprod 33:1973–1984. https://doi.org/10.1093/humrep/dey264
Mbango JN, Coutton C, Arnoult C et al (2019) Genetic causes of male infertility: snapshot on morphological abnormalities of the sperm flagellum. Basic Clin Androl 9:2. https://doi.org/10.1186/s12610-019-0083-9
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110.20
McLaren W, Gil L, Hunt SE et al (2016) The ensembl variant effect predictor. Genome Biol 17:1–14. https://doi.org/10.1186/s13059-016-0974-4
Nagirnaja L, Aston KI, Conrad DF (2018) Genetic intersection of male infertility and cancer. Fertil Steril 109:20–26. https://doi.org/10.1016/j.fertnstert.2017.10.028
Nieschlag E, Behre HM, Nieschlag S (2010) Andrology-male reproductive health and dysfunction. Springer, Berlin
Okutman O, Muller J, Baert Y et al (2015) Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum Mol Genet 24:5581–5588. https://doi.org/10.1093/hmg/ddv290
Oud MS, Volozonoka L, Smits RM et al (2019) A systematic review and standardized clinical validity assessment of male infertility genes. Hum Reprod 34:932–941. https://doi.org/10.1093/humrep/dez022
Pedersen BS, Quinlan AR (2017) Who’s who? Detecting and resolving sample anomalies in human DNA sequencing studies with peddy. Am J Hum Genet 100:406–413. https://doi.org/10.1016/j.ajhg.2017.01.017
Schlatt S, Pohl EHV et al (2019) Ageing in men with normal spermatogenesis alters spermatogonial dynamics and nuclear morphology in Sertoli cells. Andrology 7:827–839. https://doi.org/10.1111/andr.12665
Scott EM, Halees A, Itan Y et al (2016) Characterization of greater middle eastern genetic variation for enhanced disease gene discovery. Nat Genet 48:1071–1079. https://doi.org/10.1038/ng.3592
Scrucca L, Fop M, Murphy T, Raftery A (2016) mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R J 8:289–317. https://doi.org/10.1016/j.physbeh.2017.03.040
Shibuya H, Ishiguro KI, Watanabe Y (2014) The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat Cell Biol 16:145–156. https://doi.org/10.1038/ncb2896
Shibuya H, Hernández-Hernández A, Morimoto A et al (2015) MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell 163:1252–1266. https://doi.org/10.1016/j.cell.2015.10.030
Tiepolo L, Zuffardi O (1976) Localization of factors controlling spermatogenesis in the nonfluorescent portion oi the human Y chromosome long arm. Hum Genet 124:119–124. https://doi.org/10.1007/bf00278879
Tüttelmann F, Ruckert C, Röpke A (2018) Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Medizinische Genet 30:12–20. https://doi.org/10.1007/s11825-018-0181-7
Ventimiglia E, Montorsi F, Salonia A (2016) Comorbidities and male infertility: a worrisome picture. Curr Opin Urol 26:146–151. https://doi.org/10.1097/MOU.0000000000000259
Vockel M, Riera-Escamilla A, Tüttelmann F, Krausz C (2019) The X chromosome and male infertility. Hum Genet. https://doi.org/10.1007/s00439-019-02101-w
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:1–7. https://doi.org/10.1093/nar/gkq603
Wang Y, Chen Y, Chen J et al (2019) The meiotic TERB1-TERB2-MAJIN complex tethers telomeres to the nuclear envelope. Nat Commun 10:1–19. https://doi.org/10.1038/s41467-019-08437-1
Weinbauer GF, Behr R, Bergmann M, Nieschlag E (1998) Testicular cAMP responsive element modulator (CREM) protein is expressed in round spermatids but is absent or reduced in men with round spermatid maturation arrest. Mol Hum Reprod 4:9–15. https://doi.org/10.1093/molehr/4.1.9
Wilfert AB, Chao KR, Kaushal M et al (2016) Genome-wide significance testing of variation from single case exomes. Nat Genet 48:1455–1461. https://doi.org/10.1038/ng.3697
Xavier M, Salas-Huetos A, Oud M et al (2020) Disease gene discovery in male infertility: past, present and future. Hum Genet. https://doi.org/10.1007/s00439-020-02202-x
Zhang J, Tu Z, Watanabe Y, Shibuya H (2017) Distinct TERB1 domains regulate different protein interactions in meiotic telomere movement. Cell Rep 21:1715–1726. https://doi.org/10.1016/j.celrep.2017.10.061
Acknowledgements
We thank all the participants and their families for their enthusiastic collaboration. Variant calling and interpretation for Family 1 were performed at the Utah Center for Genetic Discovery Core, part of the Health Sciences Center Cores at University of Utah. We are grateful to Jochen Wistuba for help with the CREM staining of case M1646, and we thank Nadja Rotte for her help with testicular histology in controls. This work was supported in part by funding from the National Institutes of Health (R01HD078641) and the German Research Foundation (Clinical Research Unit ‘Male Germ Cells: from Genes to Function’, DFG CRU326). GEMINI Consortium members: Donald F. Conrad, Kenneth I. Aston, Douglas T. Carrell, James M. Hotaling, Liina Nagirnaja, Timothy G. Jenkins, Moira K. O’Bryan, Rob McLachlan, Peter N. Schlegel, Michael L. Eisenberg, Jay I. Sandlow, James F. Smith, Puneet Kamal, Carole Ober, Mark Sigman, Kathleen Hwang, Emily S. Jungheim, Kenan R. Omurtag, Alexandra M. Lopes, Filipa Carvalho, Susana Fernandes, Alberto Barros, João Gonçalves, Maris Laan, Margus Punab, Ewa Rajpert-De Meyts, Niels Jørgensen, Kristian Almstrup, Csilla G. Krausz, Keith A. Jarvi, Davor Jezek.
Author information
Authors and Affiliations
Consortia
Contributions
All authors contributed to the study conception and design. Web resources: Gene Cards, https://www.genecards.org/. UCSC Genome Browser, https://genome.ucsc.edu/. Genome Aggregation Database, https://gnomad.broadinstitute.org/. Greater Middle East Variome, https://igm.ucsd.edu/gme/. OMIM, https://www.omim.org/. Protein Atlas, https://www.proteinatlas.org/. PubMed, https://www.ncbi.nlm.nih.gov/PubMed/. UCSC In-Silico PCR, https://genome.ucsc.edu/cgi-bin/hgPcr. UniProt, https://www.uniprot.org/.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no competing interests.
Data and code availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2020_2236_MOESM1_ESM.tif
Fig. S1. Protein, genetic context, validation and alignment of the TERB2 variants detected in Family 1. a) Position of TERB2 on chromosome 15. b) Sequence length and exon structure for TERB2. c) Corresponding protein domain structure. d) Primers for the region of interest in exon 6, wild type sequence and validation of exon 6 TERB2 variant using Sanger sequencing in all samples analyzed. e) Primers for the region of interest in exon 7, wild type sequence and validation of exon 7 TERB2 variant using Sanger sequencing in all the analyzed samples. (TIF 1015 KB)
439_2020_2236_MOESM2_ESM.tif
Fig. S2. Protein sequence conservation of the exon 6 TERB2 variant (p.Thr153fs*17) detected in Family 1 was assessed across 100 vertebrate species using Multiz alignment in the UCSC Genome Browser. * Coelacanth was excluded from the species count because its sequence is truncated. (TIF 183 KB)
439_2020_2236_MOESM3_ESM.tif
Fig. S3. Protein sequence conservation of the exon 7 TERB2 variant (p.Met182fs*31) detected in Family 1 was assessed across 100 vertebrate species using Multiz alignment in the UCSC Genome Browser. * Coelacanth was excluded from the species count because its sequence is truncated. (TIF 231 KB)
439_2020_2236_MOESM4_ESM.tif
Fig. S4. Protein, genetic context, validation and alignment of the TERB1 variants detected in Individuals 2 and 3. a) Position of TERB1 on chromosome 16. b) Sequence length and exon structure for TERB1. c) Corresponding protein domain structure. d) Primers for the region of interest and validation of exon 11 TERB1 variant using Sanger sequencing in individual 2. e) Primers for the region of interest and validation of exon 18 TERB1 variant using Sanger sequencing in individual 3 (M2073). (TIF 591 KB)
439_2020_2236_MOESM5_ESM.tif
Fig. S5. Protein sequence conservation of the TERB1 variant (c.977A>G, p.Glu326Gly) detected in Individual 2 was assessed across 100 vertebrate species using Multiz alignment in the UCSC Genome Browser. (TIF 74 KB)
439_2020_2236_MOESM6_ESM.tif
Fig. S6. Protein, genetic context, validation and alignment of the MAJIN variant detected in Individual 4 (M1646). a) Position of MAJIN on chromosome 11. b) Sequence length and exon structure for MAJIN gene. c) Corresponding protein domain structure. d) Primers for the region of interest and validation of exon 5 MAJIN variant using Sanger sequencing in the analyzed sample. (TIF 453 KB)
439_2020_2236_MOESM7_ESM.tif
Fig. S7. Protein sequence conservation of the MAJIN variant (c.158G>A, p.Arg53His) detected in Individual 4 (M1646) was assessed across 100 vertebrate species using Multiz alignment in the UCSC Genome Browser. (TIF 73 KB)
439_2020_2236_MOESM8_ESM.tif
Fig. S8. RNA expression of TERB2, TERB1 and MAJIN genes with NX data (Consensus Normalized eXpression) levels for 55 tissue types and 6 blood cell types, showing strong enrichment in the testis, created by combining the data from the three transcriptomic datasets (HPA, GTEx and FANTOM5). (TIF 653 KB)
439_2020_2236_MOESM9_ESM.xlsx
Table S1. List of 170 candidate genes that were previously reported to be associated with impaired spermatogenesis according to a systematic review by Oud et al. 2019 and eight recently published genes associated with non-obstructive azoospermia (Krausz et al. 2020) (XLSX 13 KB)
439_2020_2236_MOESM11_ESM.xlsx
able S3 Filtered PSAP output for three patients with TERB1 and MAJIN variants based on the following criteria: 1) minor allele frequency [MAF] <0.01 in gnomAD, 1000 Genomes, NHLBI Exome Sequencing Project and ExAC databases, 2) CADD-score ≥20) and 3) PSAP p-value ≤ 0.001. Tabs 1-3 represent the highest-ranked variants for Individual 2, M2073 and M1646, respectively, and tab 4 includes the legend to define column headers in PSAP tables (XLSX 37 KB)
Rights and permissions
About this article
Cite this article
Salas-Huetos, A., Tüttelmann, F., Wyrwoll, M.J. et al. Disruption of human meiotic telomere complex genes TERB1, TERB2 and MAJIN in men with non-obstructive azoospermia. Hum Genet 140, 217–227 (2021). https://doi.org/10.1007/s00439-020-02236-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-020-02236-1