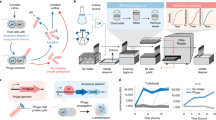
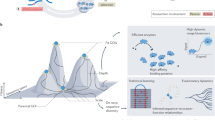
Abstract
Directed evolution, which applies the principles of Darwinian evolution to a laboratory setting, is a powerful strategy for generating biomolecules with diverse and tailored properties. This technique can be implemented in a highly efficient manner using continuous evolution, which enables the steps of directed evolution to proceed seamlessly over many successive generations with minimal researcher intervention. Phage-assisted continuous evolution (PACE) enables continuous directed evolution in bacteria by mapping the steps of Darwinian evolution onto the bacteriophage life cycle and allows directed evolution to occur on much faster timescales compared to conventional methods. This protocol provides detailed instructions on evolving proteins using PACE and phage-assisted non-continuous evolution (PANCE) and includes information on the preparation of selection phage and host cells, the assembly of a continuous flow apparatus and the performance and analysis of evolution experiments. This protocol can be performed in as little as 2 weeks to complete more than 100 rounds of evolution (complete cycles of mutation, selection and replication) in a single PACE experiment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The authors declare that any data discussed in this protocol are available in the supporting primary research papers.
References
Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 (2015).
Badran, A. H. & Liu, D. R. In vivo continuous directed evolution. Curr. Opin. Chem. Biol. 24, 1–10 (2015).
Mills, D. R., Peterson, R. L. & Spiegelman, S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc. Natl Acad. Sci. USA 58, 217–224 (1967).
Wright, M. C. & Joyce, G. F. Continuous in vitro evolution of catalytic function. Science 276, 614–617 (1997).
Esvelt, K. M., Carlson, J. C. & Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 (2011).
Berman, C. M. et al. An adaptable platform for directed evolution in human cells. J. Am. Chem. Soc. 140, 18093–18103 (2018).
Badran, A. H. et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 533, 58–63 (2016).
Badran, A. H. & Liu, D. R. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat. Commun. 6, 8425 (2015).
Barbas, C. F. Phage Display: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2001).
Carlson, J. C., Badran, A. H., Guggiana-Nilo, D. A. & Liu, D. R. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat. Chem. Biol. 10, 216–222 (2014).
Bennett, N. J. & Rakonjac, J. Unlocking of the filamentous bacteriophage virion during infection is mediated by the C domain of pIII. J. Mol. Biol. 356, 266–273 (2006).
Hubbard, B. P. et al. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat. Methods 12, 939–942 (2015).
Leconte, A. M. et al. A population-based experimental model for protein evolution: effects of mutation rate and selection stringency on evolutionary outcomes. Biochemistry 52, 1490–1499 (2013).
Packer, M. S., Rees, H. A. & Liu, D. R. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat. Commun. 8, 956 (2017).
Thuronyi, B. W. et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 37, 1070–1079 (2019).
Wang, T., Badran, A. H., Huang, T. P. & Liu, D. R. Continuous directed evolution of proteins with improved soluble expression. Nat. Chem. Biol. 14, 972–980 (2018).
Bryson, D. I. et al. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 13, 1253–1260 (2017).
Roth, T. B., Woolston, B. M., Stephanopoulos, G. & Liu, D. R. Phage-assisted evolution of Bacillus methanolicus methanol dehydrogenase 2. ACS Synth. Biol. 8, 796–806 (2019).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Suzuki, T. et al. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 13, 1261–1266 (2017).
Miller, S. M. et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 38, 471–481 (2020).
Dickinson, B. C., Leconte, A. M., Allen, B., Esvelt, K. M. & Liu, D. R. Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution. Proc. Natl Acad. Sci. USA 110, 9007–9012 (2013).
Brodel, A. K., Jaramillo, A. & Isalan, M. Engineering orthogonal dual transcription factors for multi-input synthetic promoters. Nat. Commun. 7, 13858 (2016).
Pu, J., Zinkus-Boltz, J. & Dickinson, B. C. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol. 13, 432–438 (2017).
Zinkus-Boltz, J., DeValk, C. & Dickinson, B. C. A phage-assisted continuous selection approach for deep mutational scanning of protein–protein interactions. ACS Chem. Biol. 14, 2757–2767 (2019).
Pu, J., Disare, M. & Dickinson, B. C. Evolution of C-terminal modification tolerance in full-length and split T7 RNA polymerase biosensors. Chembiochem 20, 1547–1553 (2019).
Dickinson, B. C., Packer, M. S., Badran, A. H. & Liu, D. R. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Nat. Commun. 5, 5352 (2014).
Richter, M. F. Z. et al. Phage-assisted evolution of an adenine base editor with enhanced Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020).
Brodel, A. K., Jaramillo, A. & Isalan, M. Intracellular directed evolution of proteins from combinatorial libraries based on conditional phage replication. Nat. Protoc. 12, 1830–1843 (2017).
Javanpour, A. A. & Liu, C. C. Genetic compatibility and extensibility of orthogonal replication. ACS Synth. Biol. 8, 1249–1256 (2019).
Ravikumar, A., Arzumanyan, G. A., Obadi, M. K. A., Javanpour, A. A. & Liu, C. C. Scalable, continuous evolution of genes at mutation rates above genomic error thresholds. Cell 175, 1946–1957 (2018).
Zhong, Z., Ravikumar, A. & Liu, C. C. Tunable expression systems for orthogonal DNA replication. ACS Synth. Biol. 7, 2930–2934 (2018).
Zhong, Z. et al. Automated continuous evolution of proteins in vivo. ACS Synth. Biol. 9, 1270–1276 (2020).
English, J. G. et al. VEGAS as a platform for facile directed evolution in mammalian cells. Cell 178, 1030 (2019).
Barbas, C. F. 3rd Recent advances in phage display. Curr. Opin. Biotechnol. 4, 526–530 (1993).
Ringquist, S. et al. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 6, 1219–1229 (1992).
Acknowledgements
We thank K. Esvelt, J. Carlson, B. Dickinson, A. Badran, B. Hubbard, M. Packer, D. Bryson, J. Hu, T. Roth, B. Thuronyi, M. Richter and K. Zhao for their contributions to the development of PACE and T. Blum for helpful discussion. S.M.M. was supported by a National Science Foundation Graduate Research Fellowship. T.W. was supported by a Ruth L. Kirchstein National Research Service Awards Postdoctoral Fellowship (F32GM119228). We are grateful for support from US NIH R01 EB027793, U01 AI142756, RM1 HG009490, R35 GM118062, the Howard Hughes Medical Institute and the Bill & Melinda Gates Foundation.
Author information
Authors and Affiliations
Contributions
S.M.M., T.W. and D.R.L wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.M.M., T.W. and D.R.L. have filed patent applications on PACE technologies and PACE-evolved proteins.
Additional information
Peer review information Nature Protocols thanks Mark Isalan, Chang Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Esvelt, K. M., Carlson, J. C. & Liu, D. R. Nature 472, 499−503 (2011): https://doi.org/10.1038/nature09929
Carlson, J. C., Badran, A. H., Guggiana-Nilo, D. A. & Liu, D. R. Nat. Chem. Biol. 10, 216−222 (2014): https://doi.org/10.1038/nchembio.1453
Badran, A. H. et al. Nature 533, 58−63 (2016): https://doi.org/10.1038/nature17938
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Data 1
GenBank-formatted plasmid map depicting ∆gIII selection phage with a rpoz-dSpCas9 insert as the POI. This insert can be swapped for any POI using any standard cloning method.
Rights and permissions
About this article
Cite this article
Miller, S.M., Wang, T. & Liu, D.R. Phage-assisted continuous and non-continuous evolution. Nat Protoc 15, 4101–4127 (2020). https://doi.org/10.1038/s41596-020-00410-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00410-3
This article is cited by
-
Continuous directed evolution of a compact CjCas9 variant with broad PAM compatibility
Nature Chemical Biology (2024)
-
Phage-assisted evolution of compact Cas9 variants targeting a simple NNG PAM
Nature Chemical Biology (2024)
-
Phage-assisted evolution of highly active cytosine base editors with enhanced selectivity and minimal sequence context preference
Nature Communications (2024)
-
Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity
Nature Biotechnology (2023)
-
High-throughput continuous evolution of compact Cas9 variants targeting single-nucleotide-pyrimidine PAMs
Nature Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.