Abstract
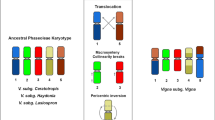
Polyploidy and dysploidy have been reported as the main events in karyotype evolution of plants. In the genus Phaseolus L. (2n = 22), a small monophyletic group of three species, the Leptostachyus group, presents a dysploid karyotype with 2n = 20. It was shown in Phaseolus leptostachyus that the dysploidy was caused by a nested chromosome fusion (NCF) accompanied by several translocations, suggesting a high rate of karyotype evolution in the group. To verify if this karyotype restructuring was a single event or occurred progressively during the evolution of this group, we analysed P. macvaughii, sister to Phaseolus micranthus + P. leptostachyus. Twenty-four genomic clones of P. vulgaris previously mapped on P. leptostachyus, in addition to the 5S and 35S rDNA probes, were used for fluorescence in situ hybridization. Only a single rearrangement was common to the two species: the nested chromosome fusion (NCF) involving chromosomes 10 and 11. The translocation of chromosome 2 is not the same found in P. leptostachyus, and pericentric inversions in chromosomed 3 and 4 were exclusive of P. macvaughii. The other rearrangements observed in P. leptostachyus were not shared with this species, suggesting that they occurred after the separation of these lineages. The presence of private rearrangements indicates a progressive accumulation of karyotype changes in the Leptostachyus group instead of an instant genome-wide repatterning.




Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- Chr:
-
Chromosome
- DAPI:
-
4,6-Diamidino-2-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- NCF:
-
Nested Chromosome Fusion
- Mya:
-
Million years ago
- rDNA:
-
Ribosomal DNA
- Ple :
-
Phaseolus leptostachyus
- Pma :
-
Phaseolus macvaughii
- Pvu :
-
Phaseolus vulgaris
References
Bonifácio EM, Fonsêca A, Almeida C, dos Santos KGB, Pedrosa-Harand A (2012) Comparative cytogenetic mapping between the lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.). Theor Appl Genet 124:1513–1520. https://doi.org/10.1007/s00122-012-1806-x
Broughton WJ, Hérnadez G, Blair MW, Beebe SE, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.)—model food legumes. Plant Soil 252:55–128. https://doi.org/10.1023/A:1024146710611
Capilla L, Sánchez-Guillén RA, Farré M, Paytuví-Gallart A, Malinverni R, Ventura J, Larkin DM, Ruiz-Herrera A (2016) Mammalian comparative genomics reveals genetic and epigenetic features associated with genome reshuffling in Rodentia. Genome Biol Evol 8:3703–3717. https://doi.org/10.1093/gbe/evw276
Carvalho CR, Saraiva LS (1993) An air drying technique for maize chromosomes without enzymatic maceration. Biotech Histochem 68:142–145. https://doi.org/10.3109/10520299309104684
Cusimano N, Souza A, Renner SS (2012) Maximum likelihood inference implies a high, not a low, ancestral haploid chromosome number in Araceae, with a critique of the bias introduced by ‘x’. Ann Bot 109:681–692. https://doi.org/10.1093/aob/mcr302
de Vos JM, Hannah A, Livio B, Kay L (2020) Speciation through chromosomal fusion and fission in Lepidoptera. Philos Trans R Soc B 375:20190539. https://doi.org/10.1098/rstb.2019.0539
Delgado-Salinas A, Bibler R, Lavin M (2006) Phylogeny of the genus Phaseolus (Leguminosae): a recent diversification in an ancient landscape. Syst Bot 31:779–791. https://doi.org/10.1600/036364406779695960
Escudero M, Hahn M, Brown BH, Lueders K, Hipp AL (2016) Chromosomal rearrangements in holocentric organisms lead to reproductive isolation by hybrid dysfunction: the correlation between karyotype rearrangements and germination rates in sedges. Am J Bot 103:1529–1536. https://doi.org/10.3732/ajb.1600051
Fonsêca A, Pedrosa-Harand A (2013) Karyotype stability in the genus Phaseolus evidenced by the comparative mapping of the wild species Phaseolus microcarpus. Genome 56:335–343. https://doi.org/10.1139/gen-2013-0025
Fonsêca A, Ferreira J, dos Santos TRB, Mosiolek M, Bellucci E, Kami J, Gepts P, Geffroy V, Schweizer D, dos Santos KGB, Pedrosa-Harand A (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosom Res 18:487–502. https://doi.org/10.1007/s10577-010-9129-8
Fonsêca A, Ferraz ME, Pedrosa-Harand A (2016) Speeding up chromosome evolution in Phaseolus: multiple rearrangements associated with a one-step descending dysploidy. Chromosoma 125:413–421. https://doi.org/10.1007/s00412-015-0548-3
Fuller ZL, Koury SA, Phadnis N, Schaeffer SW (2019) How chromosomal rearrangements shape adaptation and speciation: case studies in Drosophila pseudoobscura and its sibling species Drosophila persimilis. Mol Ecol 28:1283–1301. https://doi.org/10.1111/mec.14923
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885. https://doi.org/10.1093/nar/7.7.1869
Guerra M (2000) Chromosome number variation and evolution in monocots. In: Wilson KL, Morrison DA (eds) Monocots - Systematics and Evolution. CSIRO, Melbourne, pp 127–136
Guerra M (2008) Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenet Genome Res 120:339–350. https://doi.org/10.1159/000121083
Guerra M, Souza MJ (2002) Como observarcromossomos: Um Guia de Técnica sem Citogenética Vegetal, Animal e Humana. 1a edição, Ribeirão Preto, FUNPEC, pp 23
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA: DNA in situ hybridization preparations. Trends Genet 8:372–373. https://doi.org/10.1016/0168-9525(92)90287-E
Hoagland DR, Arnon DI (1950) The water culture method for growing plants without soils. Berkeley, California Agric Exp Station 347:29–32
Idziak D, Hazuka I, Poliwczak B, Wiszynska A, Wolny E, Hasterok R (2014) Insight into the karyotype evolution of Brachypodium species using comparative chromosome barcoding. PLoS One 9:e93503. https://doi.org/10.1371/journal.pone.0093503
International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768. https://doi.org/10.1038/nature08747
Khandelwal S (1990) Chromosome evolution in the genus Ophioglossum L. Bot J Linn Soc 102:205–217. https://doi.org/10.1111/j.1095-8339.1990.tb01876.x
Li J, Yuan J, Zhao Y, Meng F, Liu C, Zhang Z, Guo H, Xie Y, Hou Y, Li X, Wang X (2019) Reconstruction of evolutionary trajectories of coffee chromosomes. BMC Genomics 20:180. https://doi.org/10.1186/s12864-019-5566-8
Luo MC, Deal KR, Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, Crossman CC, Dubcovsky J, Gill BS, Gu YQ, Hadam J, Heo HY, Huo N, Lazo G, Ma Y, Matthews DE, McGuire PE, Morrell PL, Qualset CO, Renfro J, Tabanao D, Talbert LE, Tian C, Toleno DM, Warburton ML, You FM, Zhang W, Dvorak J (2009) Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA 106:15780–15785. https://doi.org/10.1073/pnas.0908195106
Lysak MA, Pecinka A, Schubert I (2003) Recent progress in chromosome painting of Arabidopsis and related species. Chromosom Res 11:195–204. https://doi.org/10.1023/A:1022879608152
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci U S A 103:5224–5229. https://doi.org/10.1073/pnas.0510791103
Mandáková T, Guo X, Özüdoğru B, Mummenhoff K, Lysak MA (2018) Hybridization-facilitated genome merger and repeated chromosome fusion after 8 million years. Plant J 96:748–760. https://doi.org/10.1111/tpj.14065
Mandáková T, Hloušková P, Windham MD, Mitchell-Olds T, Ashby K, Price B, Carman J, Lysak MA (2020) Chromosomal evolution and Apomixis in the cruciferous tribe Boechereae. Front Plant Sci 11:514. https://doi.org/10.3389/fpls.2020.00514
Mas de Xaxars G, Garnatje T, Pellicer J, Siljak-Yakovlev S, Vallès J, Garcia S (2015) Impact of dysploidy and polyploidy on the diversification of high mountain Artemisia (Asteraceae) and allies. Alp Bot 126:35–48. https://doi.org/10.1007/s00035-015-0159-x
Mercado-Ruaro P, Delgado-Salinas A (1998) Karyotypic studies on species of Phaseolus (Fabaceae: Phaseolinae). Am J Bot 85:1–9. https://doi.org/10.2307/2446547
Oliveira ARS, Martins LD, Bustamante FO, Muñoz-Amatriaín M, Close T, da Costa AF, Benko-Iseppon AM, Pedrosa-Harand A, Brasileiro-Vidal AC (2020) Breaks of macrosynteny and collinearity among moth bean (Vigna aconitifolia), cowpea (V. unguiculata), and common bean (Phaseolus vulgaris). Chromosom Res. https://doi.org/10.1007/s10577-020-09635-0
Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Pedrosa-Harand A, de Almeida CC, Mosiolek M, Blair MW, Schweizer D, Guerra M (2006) Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet 112:924–933. https://doi.org/10.1007/s00122-005-0196-8
Schmutz J, McClean PE, Mamidi S et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713. https://doi.org/10.1038/ng.3008
Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27:207–216. https://doi.org/10.1016/j.tig.2011.03.004
Schubert I, Vu GTH (2016) Genome stability and evolution: attempting a holistic view. Trends Plant Sci 21:749–757. https://doi.org/10.1016/j.tplants.2016.06.003
Udall JA, Long E, Ramaraj T, Conover JL, Yuan D, Grover CE, Gong L, Arick MA II, Masonbrink RE, Peterson DG, Wendel JF (2019) The genome sequence of Gossypioides kirkii illustrates a descending dysploidy in plants. Front Plant Sci 10:1541. https://doi.org/10.3389/fpls.2019.01541
Vanzela AL, Guerra M, Luceno M (1996) Rhynchospora tenuis link (Cyperaceae), a species with the lowest number of holocentric chromosomes. Cytobios 88:219–228
Wang H, Yin H, Jiao C, Fang X, Wang G, Li G, Ni F, Li P, Su P, Ge W, Lyu Z, Xu S, Yang Y, Hao Y, Cheng X, Zhao J, Liu C, Xu F, Ma X, Sun S, Zhao Y, Bao Y, Liu C, Zhang J, Pavlicek T, Li A, Yang Z, Nevo E, Kong L (2020) Sympatric speciation of wild emmer wheat driven by ecology and chromosomal rearrangements. PNAS 117:5955–5963. https://doi.org/10.1073/pnas.1920415117
Weising K, Nybom H, Wolff K, Kahl G (2005) CTAB protocol I. In: DNA fingerprinting in plants : principles, methods, and applications, 2nd edn. Taylor & Francis Group, pp 100–102
Winterfeld G, Ley A, Hoffmann MH, Paule J, Röser M (2020) Dysploidy and polyploidy trigger strong variation of chromosome numbers in the prayer-plant family (Marantaceae). Plant Syst Evol 306:36. https://doi.org/10.1007/s00606-020-01663-x
Yogeeswaran K, Frary A, York T, Amenta A, Lesser AH, Nasrallah JB, Tanksley SD, Nasrallah ME (2005) Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res 15:505–515. https://doi.org/10.1101/gr.3436305
Acknowledgements
The authors thank Embrapa Arroz e Feijão (Santo Antônio de Goiás, Brazil) and the International Center of Tropical Agriculture – CIAT (Cali, Colombia), for seed supply; Paul Gepts (University of California, Davis, USA) for supplying the BAC clones from P. vulgaris; and Valérie Geffroy (Université Paris-Sud, Orsay Cedex, France) for the bacteriophage SJ19.12 from P. vulgaris. The authors also thank Gustavo Souza for help with image editing, as well as Tiago Ribeiro and Thiago Henrique do Nascimento for critical reading of the manuscript. We are also thankful to the National Council for Scientific and Technological Development – CNPq and CAPES (Coordenação de Pessoal de Nível Superior: Finance Code001) Brazil, for financial support.
Funding
The present study received financial support and fellowships from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and CAPES (Coordenação de Pessoal de Nível Superior: Finance Code 001).
Author information
Authors and Affiliations
Contributions
MEF: performed experiments, organized the figures, and drafted the manuscript. AF: analysed and discussed the data. APH: designed and supervised the research and corrected the manuscript. All authors read, discussed, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors approved the final version of the manuscript.
Code availability
Not applicable.
Additional information
Responsible Editor: Jiming Jiang
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key message
After a nested chromosome fusion (NCF) and consequent dysploidy, successive and independent rearrangements differentiated the chromosome complements of two species of the Leptostachyus group in a relatively short period of time.
Rights and permissions
About this article
Cite this article
Ferraz, M.E., Fonsêca, A. & Pedrosa-Harand, A. Multiple and independent rearrangements revealed by comparative cytogenetic mapping in the dysploid Leptostachyus group (Phaseolus L., Leguminosae). Chromosome Res 28, 395–405 (2020). https://doi.org/10.1007/s10577-020-09644-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-020-09644-z