Abstract
Intra-species genetic variability assessment is an effective tool in formulating genetic improvement and germplasm conservation strategies. Houttuynia cordata Thunb. is a semidomesticated medicinal herb consumed widely in traditional diet in North-Eastern India. In the present study, an effort has been made to assess the genetic diversity of H. cordata Thunb. from Brahmaputra valley of North-East India. A total of 545 genotypes from 18 populations of H. cordata Thunb. from four different regions, i.e. North-East, North-West, South-East and South-West, with respect to river Brahmaputra were collected and population genetic diversity and structure were analysed using ISSR molecular markers. Population genetic structure analysis using unweighted pair group method with averages (UPGMA)-based hierarchical cluster analysis, principal coordinate analysis (PCoA) and model-based clustering in STRUCTURE program revealed that the population of H. cordata Thunb. grouped according to regional distribution and forms four genetically distinct clusters. The analysis of molecular variance showed that differentiation among regions was significant with 60% genetic variation among region, 3% genetic variation among population within region and 37% genetic variation within population. We found wide variation in Nei’s gene diversity (Hj) ranging from 0.07782 in Margherita population to 0.13634 in Barapani population. Furthermore, Nei’s gene diversity within population (Nei’s Hs) and total gene diversity (Ht) were found to be 0.1081 and 0.1769 respectively. The genetic differentiation among 18 population was high (Fst = 0.3894; p < 0.001) with relatively restricted gene flow (Nm = 0.6564). Based on the result of this study, we suggest ex situ conservation could be an appropriate measure to adequately capture the total genetic diversity of H. cordata Thunb. populations of North-East India by selecting few individuals from different populations.
Similar content being viewed by others
Introduction
Houttuynia cordata Thunb. is a perennial herbaceous medicinal plant distributed in Japan, China, Korea, and Southeast Asia and the Himalayas. In India, it is mainly distributed in North-Eastern states including Assam, Meghalaya, Manipur, Tripura, Mozoram, Arunachal Pradesh and Sikkim (Bhattacharyya and Sharma 2010). The plant prefers to grow in moist and shady habitat (Alves et al. 2013; Wei and Wu 2012). A study conducted in North-East India has indicated that the plant requires a special moist habitat having clay loam soil with higher organic carbon (0.85 ± 0.05%), average soil pH 5.9 and 78% soil moisture (Bhattacharyya and Sharma 2010). It is the only member of the genus Houttuynia under the family Saururaceae (Fang 1981; Tseng 1982). The plant is popularly known by different common names like Chameleon plant, fish wort or heart leaf, and Chinese lizard tail. It is widely used for ornamental (Frank and Dana 2012), culinary and medicinal purposes (Wu et al. 2002). Mature plants of H. cordata Thunb. are 30–60 cm tall, with horizontal rhizomes or stolons that connect the erect aerial stems. The terminal inflorescences consist of dense spikes (1.5–2.5 cm long) of very small flowers without petals, but each inflorescence has four bracts at the base which resembles a single flower. The leaves are heart-shaped, cordate with a sheathing base, measuring up to 10 cm long and 6 cm wide, usually green but can turn into purplish colour when exposed to sunlight. The fruits are tiny (2–3 mm long) with 9 to 18 apomictic seeds in each (Alves et al. 2013). Sexual reproduction has not been reported for this plant (Wei and Wu 2012). It mainly reproduces through vegetative propagation by producing abundant rhizomes or stolons and by parthenogenesis (Mihara 1960; Shibata and Miyake 1908).
All parts of the plants are edible with high medicinal properties and have been used traditionally for curing various human diseases (Bhattacharyya and Sharma 2010). Different studies indicate that H. cordata Thunb. contains abundant nutrients and active components including volatile oils, flavonoids and water-soluble polysaccharides (Li et al. 2005; Cao and Wang 2005). In addition, H. cordata Thunb. exhibits a wide range of pharmaceutical activities including antibacterial (Lu et al. 2006a; Kim et al. 2008), antiviral (Hayashi et al. 1995; Liu et al. 2010; Chen et al. 2011; Yin et al. 2011), anti-inflammatory (Lu et al. 2006b; Shin et al. 2010; Li et al. 2011; Kim et al. 2012), immunologic (Lau et al. 2008), anticancer (Zhou et al. 2012; Lai et al. 2010), antioxidative (Chen et al. 2003; Toda 2005) and anti-mutagenic (Chen et al. 2004) effects. The extract of H. cordata Thunb. has been demonstrated to have immunomodulatory and anti-SARS (severe acute respiratory syndrome) activities (Lu et al. 2006b; Lau et al. 2008). Also, it was reported to inhibit human immunodeficiency virus type 1 (HIV-1) and herpes simplex virus type 1 (HSV-1) (Hayashi et al. 1995). In North-East India, it is widely used by local tribes as ethno medicine (Sarkar 1993; Bora 2001; Khongsai et al. 2011). It is eaten raw by local tribal people for lowering blood sugar level (Marles and Farnsworth 1995). The juices of leaf are traditionally used for treatment of diseases like cholera, dysentery and anaemia and purification of blood (Laloo and Hemalatha 2011).
There is no record of organized cultivation of this medicinal plant, and therefore, it is mostly harvested from its wild habitat. With the growth of commercial demand in the recent years, the plant is being indiscriminately harvested from wild resulting in habitat destruction, population decrease and reduction in its genetic diversity (Zhong et al. 2009). Many studies from China have reported genetic diversity of H. cordata Thunb. based on morphological features, isozyme, chromosome number and ISSR markers (Wu et al. 2002, 2003; Wei and Wu 2012; Zhong et al. 2016). But until now, no study has targeted the genetic diversity and population structure of this important medicinal plant in North-East India, although this information is essential for the formulation of any effective conservation strategies. In the present work, 18 populations of H. cordata Thunb. from different regions of North-East India were analysed using ISSR marker to study their genetic diversity, differentiation and structure. We then used these results to describe genetic diversity and population structure and to infer whether any gene flow among these populations occurs. The information generated in the present study on population genetics of H. cordata Thunb. could be used to plan adequate conservation actions.
Materials and Methods
Population Sampling and DNA Extraction
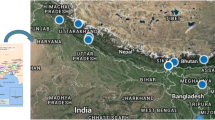
In the present study, 18 populations of H. cordata Thunb. (28–32 individuals per population) were sampled from 18 locations of North-East India during 2012 and 2013 (Fig. 1, Table 1). All these collection sites are largely distributed on Northern and Southern bank of river Brahmaputra. We divided our sampling areas into four regions with respect to the river Brahmaputra. These regions are (1) North-Eastern region which includes Nalbari (NB), Dhemaji (DM), Itanagar (IN), Kokrajhar (KJ) and North Lakhimpur (NL); (2) South-Western region which includes Cherapunji (CP), Shora Shella (SS), Mawlai (ML), Barapani (BP) and Laitryngew (LG); (3) South-Eastern region which includes Nagaon (NG), Margherita (MR), Golaghat (GG), Tinsukia (TS), Guwahati (GW) and Jeypore (JP) and (4) North-Western region which includes Kalimpong (KP) and Gangtok (GT). From each of the sampling site, random sampling was done covering an area of about 2-km radius. While collecting the leaf samples from each of the population, care was taken to maintain a minimum distance of at 0.3 km among sampled individuals to avoid sampling clonal relatives. Young leaves for DNA extraction were collected in zip-lock polybags, temporarily stored on ice until brought to the laboratory for storage at − 80 °C. The DNA from leaf was extracted following the procedure described by Doyle (1991).
Distribution of the field sampling sites in North-East India. Sampling area was divided into four regions: North-Western; North-Eastern; South-Western; South-Eastern, with respect to river Brahmaputra. The pie diagrams on the left shows the results of AMOVA where AP = among populations, WP = within populations, AR = among regions. Full name of the populations is given in Table 1
ISSR Fingerprinting
ISSR fingerprinting was done according to Wu et al. (2005), with minor modifications. Twenty-two ISSR primers were screened against 18 populations, taking two individuals randomly from each population. The PCR reaction was set in a 10 μl reaction volume using 20 ng of template DNA. The PCR reaction mixture contains 10× reaction buffer—1 μl, 2.5 mM dNTPs—0.6 μl, 10 mM primers—2 μl, template DNA—20 ng, Taq DNA polymerase (1 U/μl)—0.5 μl and nuclease-free water—to make the final volume 10 μl. The PCR profile used include initial denaturation at 94 °C for 3 min followed by 35 cycles of 94 °C for 1 min, 50 °C for 1 min and 72 °C for 2 min and a final extension of 72° for 10 min. After PCR, the amplified products were resolved in 1.3% agarose gel and photograph was taken in a gel doc for scoring. Low Range DNA Ruler Plus (Genie, Cat. No. 2662770501730) was used to determine the fragments sizes for scoring. To create a binary matrix, amplified fragments of 100 to 1200 bp were scored visually from printed gel photos as having present (1), absent (0), or ambiguous (?). Only distinct bands were scored as present, and the manual scoring procedure was repeated twice on separate occasions to reduce scoring errors.
Genetic Data Analysis
In the present study, all the genetic diversity and differentiation statistics were computed using AFLP-SURV v 1.0 program (Vekemans 2002; Vekemans et al. 2002). Bayesian method with non-uniform prior distribution of allele frequencies was used to estimate band frequency distribution (Zhivotovsky 1999) followed by genetic diversity analysis within and between population (Lynch and Milligan 1994). Banding pattern across population, proportion of polymorphic loci (PPL) at 5% level, Nei’s gene diversity (Hj), Shannon diversity index (I), Wright fixation index, Fst (Weir and Cockerham 1984), population differentiation (Gst) and gene flow (Nm = 0.5(1 − Gst)/Gst) were also computed in GenAlEx V6.5 (Peakall and Smouse 2006, 2012) and POPGENE v1.32 (Yeh et al. 1997).
To differentiate among populations, AMOVA was computed in GenAlEx v6.5 using 9999 permutations. The UPGMA (Sokal and Michener 1958) method was used for cluster analysis among populations. Nei’s minimum distance matrix (Nei 1972, 1978) as obtained from AFLP-SURV v1.0 was used for computing UPGMA clustering analysis with 10,000 bootstrap analysis of cluster relationships using NEIGHBOR module in the PHYLIP v 3.695 (Felsenstein 2008) software package.
Principal coordinate analysis (PCoA) was performed based on Nei’s minimum distance matrix (Nei 1978) in GenAlEx v 6.5. Population structure was determined using the program STRUCTURE v 2.3.3 (Falush et al. 2007) which statistically determines the probability of an individual to be included in a homogenous cluster (K populations) without prior population information. The correlated band frequencies and admixed model were applied with a burn-in-period of 100,000 and 1,000,000 MCMC replicates after burn-in. The range of clusters (K) was given from 1 to 18. From K = 1 to K = 10, ten runs were performed, whereas for K > 10, five runs were performed. The detection of the most-likely K was done according to Evanno et al. (2005), which uses LnP(D) to get information about most probable number of groups and calculate height of the modal value of the ∆K distribution to detect the most-likely K (Evanno et al. 2005).
Results
Understanding of the genetic diversity and population structure is very much essential to plan any conservation or genetic improvement program for any species. A total of ten best ISSR primers (S1 Table) that showed reproducible polymorphic amplification during screening step were used to analyse 545 individual genotypes of H. cordata Thunb. covering 18 populations, resulted in 137 markers. The number of bands amplified ranged from 28 to 47 with an average of 40.7 bands per population. The band pattern across these 18 populations is given in supplementary information 2 and Fig. 2.
Total band patterns across populations of H. cordata. no. Bands, number of different bands; No. Bands Freq. >=5%, number of different bands with a frequency ≥ 5%; no. Private Bands, number of bands unique to a single population; no. LComm Bands (<=25%), number of locally common bands (Freq. ≥ 5%) found in 25% or fewer populations; no. LComm Bands (<=50%), number of locally common bands (Freq. ≥ 50%) found in 50% or fewer populations; He, expected gene diversity; uHe, unbiased expected gene diversity. Full name of the populations is given in Table 1
Population Genetic Structure
Hierarchical cluster analysis (UPGMA) revealed that populations sampled from South-West, North-West and North-East regions of Brahmaputra valley were grouped separately (Fig. 3). Cluster I, with a bootstrap value of 100%, included all the populations of South-West region of Bramaputra valley. Cluster I can be divided into two groups, viz., group I contains two populations (CP and LG), whereas group II contains the remaining 3 populations (ML, BP, SS). Cluster II, with a bootstrap value of 100%, included all the populations sampled from North-East region of the Brahmaputra valley. Cluster III, with a bootstrap value of 100%, contained the two populations (GT and KP) sampled from North-Western region of the valley. The UPGMA clustering also revealed that all the populations from Northern Region were well separated from the populations of Southern region of Brahmaputra valley. The remaining South-Eastern populations could not be grouped into a single cluster where population MR was found to be grouped with NG. The principal coordinate analysis also revealed that populations of H. cordata Thunb. grouped according to regional distribution with Northern populations (KJ, NB, DM, IN and NL) well separated from Southern populations (BP, ML, CP, LG and SS) (Fig. 4), which is consistent with the results of UPGMA-based cluster analysis. Furthermore, our model-based clustering analysis using program STRUCTURE (Pritchard et al. 2000; Falush et al. 2003; Evanno et al. 2005) showed likelihood of LnP(D) was greatest when K = 4. We found that for K > 4, LnP(D) increased slightly but more or less plateaued (Fig. 5a), i.e. ∆K reached its maximum at K = 4 (Fig. 5b), thus indicating that all our selected populations can be included into one of the four clusters, which interestingly corresponds to the geographic distribution of these populations (Fig. 1).
UPGMA tree of 18 H. cordata populations (numbers indicate bootstrap support values). Full name of the populations is given in Table 1
Principal coordinate analysis of 18 H. cordata populations, based on Nei’s unbiased genetic distances. Horizontal and vertical scales represent the first and second principal axes of variation respectively (the two axes that explain the most variations among populations). Here the first principal axis represents a large 60.61% of variation, the second axis represents 23.59% of variation. Full name of the populations is given in Table 1
STRUCTURE analysis of H. cordata populations based on ISSR data (a the relationship between K and mean L(K); b the relationship between K and ∆K; c the grouping when K = 4). Full name of the populations is given in Table 1
The analysis of molecular variance indicated that majority of genetic variation (60%) occurred among regions, while variations among population within region and within population were found to be 3% and 37% respectively (Fig. 1). Interestingly, AMOVA showed high genetic variation within population (91%) than that of among populations (9%) when the South-Eastern population was treated as one group. Also, similar type of result was observed for the North-Western, North-Eastern and South-Western populations (Table 2), which may be one of the reasons for populations getting clustered together based on their regions. When the North-Western and North-Eastern populations were pooled together, AMOVA calculated higher genetic variation within population (56%), while variation among population was 44%. Similar types of results were also obtained when South-Western and South-Eastern populations were pooled together with variation within population (53%) more than variations among populations (47%). Moreover, when we pooled all the 18 populations together as one group and estimated variation among and within population, we found Fst value to be 0.575 (P < 0.001) with 57% variation among populations and 43% within populations. This finding thus suggests that inter-population variation was comparatively more than intra-population variation. Therefore, we hypothesise that the differentiation among regions in H. cordata Thunb. population was significant, but genetic variation within population was also maintained.
Population Genetic Diversity
For each population, the data for number of loci scored, number as well as proportion of polymorphic loci at 5% level, Nei’s gene diversity (Hj) etc. are given in supplementary information 3. The Nei’s gene diversity (Hj) ranged from 0.07782 (PLP = 21.2%) in population MR to 0.13634 (PLP = 32.1%) in population BP. Thus, high variability was observed in all populations collected from South-West region whereas low variability was recorded in population from South-East region of Brahmaputra valley. The mean Nei’s gene diversity within population (or Nei Hs) was found to be 0.1081 (supplementary information 4). Moreover, total gene diversity (Ht), i.e. gene diversity among population, was found to be 0.1769 (supplementary information 5). The average gene diversity within population (Hw) and average gene diversity among population (Hb) or genetic differentiation among population were found to be 0.1081 and 0.0689 respectively. Furthermore, Wright fixation index (Fst) showing the proportion of the total gene diversity that occurs among as opposed to within population was found to be 0.3894. When the populations were divided into four clusters based on STRUCTURE analysis (supplementary information 6), Nei’s gene diversity (Hj) was found to be 0.0776, 0.1199, 0.1057 and 0.1257 respectively for South-Eastern, North-Western, North-Eastern and South-Western populations. Similarly, Shannon diversity index (I value) for the four clusters was found to be 0.1174, 0.1866, 0.1588 and 0.1829 respectively for South-Eastern, North-Western, North-Eastern and South-Western populations. When North-Western and North-Eastern populations were pooled together, Nei’ gene diversity (Hj) was found to be 0.1492 with the Shannon diversity index value of 0.2393. Furthermore, when South-Eastern and South-Western populations were pooled together, Nei’s gene diversity (Hj) was found to be 0.1296 with the Shannon diversity index value of 0.2049.
Population Genetic Distance and Gene Flow
The 18 populations of H. cordata Thunb. in the present study showed high and significant genetic differentiation (Fst = 0.3894, P < 0.001; Gst = 0.4324). However, the estimated gene flow, Nm (Nm = 0.5(1 − Gst)/Gst), was found to be 0.6564 which suggests that although the genetic differentiation among studied populations is significant, there may be low migration rate among populations. We also calculated genetic differentiation and gene flow for the four groups separately (based on UPGMA tree, PCoA and STRUCTURE analysis results) and found that Gst, among all South-Eastern populations, was 0.0715 and Nm was 6.4974. Similarly, Gst among North-Western population was 0.0825 and Nm was 5.5591. The Gst among North-Eastern and South-Western populations was 0.0764 and 0.0586 with Nm value of 6.0474 and 8.0384 respectively. These results indicate weak genetic differentiation and frequent gene flow within the genotypes of all the regions. When the North-Western and North-Eastern populations were pooled together, the Gst value among population was 0.3263 and Nm was 1.0324 which indicates that there is significant genetic differentiation among the populations of North-Eastern and North-Western regions and that the gene flow is restricted. We also pooled the South-Eastern and South-Western populations together and calculated their respective Gst and Nm values. Interestingly, we found Gst to be 0.2868 and Nm value was 1.2433. Thus, gene flow was slightly more among South-Eastern and South-Western populations compared to between North-Eastern and North-Western populations.
Discussion
Population Genetic Diversity
In the present study, ISSR-based analysis of H. cordata Thunb. populations revealed overall high genetic diversity (Ht = 0.1769). This corroborates with results obtained by Wei and Wu (2012) in which they reported genetic variation and population differentiation of 15 populations of H. cordata Thunb. in central, South Eastern and South Western China. Generally, it is believed that genetic diversity of asexually reproducing plants is low (Hamrick and Golt 1990). However, several studies on asexually reproducing plants such as Leymus chinensis (Zu and Cui 2002), Cyclobalanopsis glauca (Chen and Song 1997) and Potamogeton malianus (Chen et al. 2006) suggest that genetic diversity of asexually reproducing plants can also be very high. Moreover, we observed comparatively high average gene diversity within population (Hw = 0.1081) than among populations (Hb = 0.0689). Similar results were also obtained by Wei and Wu (2012) using ISSR marker and Wu et al. (2002) using RAPD marker.
Population Structure and Gene Flow
The results of the hierarchical and model-based cluster analyses of ISSR data strongly suggested that the 18 sampled populations of H. cordata Thunb. could be grouped into four clusters based on the regions of sample collection—South-Western, North-Eastern, North-Western and South-Eastern regions of Brahmaputra river regardless of the approach used for genetic structure analyses. AMOVA also clearly indicated that the genetic variation within population among regions was much higher than that among populations within region. This similarity between the clustering pattern and geographical distribution of populations of H. cordata Thunb. suggests continued gene flow among populations of the same region (Schaal et al. 1998) and restricted gene flow between populations of different regions. We found high genetic differentiation but restricted gene flow between populations which were geographically fragmented (Gst = 0.4324, Nm = 0.6564) due to the river Brahmaputra. Since H. cordata Thunb. is a short, small plant and reported to propagate mostly by formation and separation of underground stems or parthenogenesis (Mihara 1960), the probability of gene flow via seed and pollen dispersal is very low. Moreover, polyploidy complex of H. cordata Thunb. with 36 to 126 chromosomes (Wu et al. 2002) will hinder gene exchanges among populations with different chromosome numbers (Wu et al. 2005). These facts suggest that the observed pattern of genetic differentiation and population structure may not be due to ongoing gene flow rather due to historical association among populations. Earlier studies have suggested that during late tertiary to early quaternary period, H. cordata Thunb. survived and adapted through asexual reproduction in the central and southern areas of the Northern hemisphere (Liang 1995). Furthermore, during glacial maxima, H. cordata Thunb. was restricted in small climatically stable refugia areas in small population size (Liang 1995), the genetic consequence of which may be high level of genetic drift (Barrett and Kohn 1991; Ellstrand and Elam 1993). This could be one reason for observed high genetic variation between regions and low genetic variation among population within region. Similar explanation was also suggested by Zhong et al. 2016, where they investigated genetic diversity and population structure of H. cordata Thunb. from China using sequence-related amplified polymorphism (SRAP) marker.
Conclusion
In conclusion, we observe that the total genetic diversity of H. cordata Thunb. was high with higher genetic differentiation between populations of different regions compared to population of the same region in North-East India. The observed genetic structure may probably be due to genetic drift of small isolated populations promoting high genetic variation between population of different regions than between population of same region. Therefore, it is important for conservation planners to devise conservation strategies for conservation of maximum possible numbers of population from all the regions rather than focusing on a few targeted populations. We, therefore, suggest to target genotypes from as many populations possible rather than few selected populations for ex situ or in situ conservation of wild populations of H. cordata Thunb. in the North-Eastern region of India.
References
Alves RJV, Medeiros D, Moura RL, Gusmao LCT, Silva NG, Antunes K, Matozinhos C, Blanco MA (2013) First report of naturalization of Houttuynia cordata Thunb. 1783 (Piperales: Saururaceae) in South America. Check List 9(3):642–644
Barrett SCH, Kohn JK (1991) Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York
Bhattacharyya N, Sharma S (2010) Assessment of availability, ecological feature, and habitat preference of the medicinal herb Houttuynia cordata Thunb. in the Brahmaputra Valley of Assam, India. Environ Monit Assess 160:277–287
Bora C (2001) Ethnobotany of lower Subansiri District (Nishi Tribe) of Arunachal Pradesh, India. Ph.D. Thesis, Gauhati University, Assam, India
Cao H, Wang SY (2005) The chemical constituent of H. cordata and the prospect of development. J Southeast Guizhou Natl Teach Coll 23:20–22
Chen XY, Song YC (1997) Clonal diversity in Cyclobalanopsis glauca populations and its relationship with environmental factors. Chin J Plan Ecol 21(4):342–348
Chen YY, Liu JF, Chen CM, Chao PY, Chang TJ (2003) A study of the antioxidative and antimutagenic effects of Houttuynia cordata Thunb. using an oxidized frying oil-fed model. J Nutr Sci Vitaminol 49:327–333
Chen L, Wu W, Zheng YL (2004) HPLC analysis on dissociative amino acids of Houttuynia. Amino Acids Biot Resour 1:20–24
Chen Y, Li Q, Wu W, Li W (2006) Genetic diversity and clonal structure of Potamogeton malaianus populations. Ying Yong Sheng Tai Xue Bao 17:2034–2040
Chen X, Wang Z, Yang Z, Wang J, Xu Y, Tan R, Li E (2011) Houttuynia cordata blocks HSV infection through inhibition of NF-kB activation. Antivir Res 92:341–345
Doyle J (1991) DNA protocols for plants. In: Hewitt GM, Johnston AWB, Young JPW (eds) Molecular techniques in taxonomy. Springer, Berlin, Heidelberg, pp 283–293
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implication for plant conservation. Annu Rev Ecol Syst 24:217–242
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genet 164(4):1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fang WP (1981) Flora Sichuanica. Sichuan People Press, Chengdu, pp 126–127
Felsenstein J (2008) PHYLIP: phylogeny inference package, version 3.68, Seattle, WA: University of Washington. Available: http://evolution.gs.washington.edu/phylip.html. Accessed on 19 Jul 2019
Frank KA, Dana MN (2012) Spreading ornamental plants: virtues & vices. Lands Hort 1:1–4
Hamrick JL and Golt MJW (1990) Allozyme diversity in plant species. In: Plant population genetics, breeding and genetic resources (Brown AHD, Clegg MJ and Kahler AL, eds.). Sinauer, Sunderland, 43–63
Hayashi K, Kamiya M, Hayashi T (1995) Virucidal effects of the steam distillate from Houttuynia cordata and its components on HSV-1, influenza virus, and HIV. Planta Med 61:237–241
Khongsai M, Saikia SP, Kayang H (2011) Ethnomedicinal plants used by different tribes of Arunachal Pradesh. Indian J Tradit Knowl 10(3):541–546
Kim GS, Kim DH, Lim JJ, Lee JJ, Han DY, Lee WM, Jung WC, Min WG, Won CG, Rhee MH, Lee HJ, Kim S (2008) Biological and antibacterial activities of the natural herb Houttuynia cordata water extract against the intracellular bacterial pathogen salmonella within the RAW 264.7 macrophage. Biol Pharm Bull 31:2012–2017
Kim D, Park D, Kyung J, Yang YH, Choi EK, Lee BY, Kim HK, Hwang BY, Kim YB (2012) Anti-inflammatory effects of Houttuynia cordata supercritical extract in carrageenan-air pouch inflammation model. Lab Anim Res 28:137–140
Lai KC, Chiu YJ, Tang YJ, Lin KL, Chiang JH (2010) Houttuynia cordata Thunb. extract inhibits cell growth and induces apoptosis in human primary colorectal cancer cells. Anticancer Res 30:3549–3556
Laloo D, Hemalatha S (2011) Ethnomedicinal plants used for diarrhea by tribals of Meghalaya, Northeast India. Pharmacogn Rev 5(10):147–154
Lau KM, Lee KM, Koon CM, Cheung CS, Lau CP et al (2008) Immunomodulatory and anti-SARS activities of Houttuynia cordata. J Ethnopharmacol 118:79–85
Li GZ, Chai OH, Lee MS, Han EH, Kim HT, Song CH (2005) Inhibitory effects of Houttuynia cordata water extracts on anaphylactic reaction and mast cell activation. Biol Pharm Bull 28:1864–1868
Li W, Zhou P, Zhang Y, He L (2011) Houttuynia cordata, a novel and selective COX-2 inhibitor with anti-inflammatory activity. J Ethnopharmacol 133:922–927
Liang HX (1995) On the evolution and distribution in Saururaceae. Acta Bot Yunnanica 17(3):255–267
Liu FZ, Shi H, Shi YJ, Liu Y, Jin YH, Gao YJ, Guo SS, Cui XL (2010) Pharmacodynamic experiment of the antivirus effect of Houttuynia cordata injection on influenza virus in mice. Acta Pharm Sin 45(3):399–402
Lu H, Wu X, Liang Y, Zhang J (2006a) Variation in chemical composition and antibacterial activities of essential oils from two species of Houttuynia cordata Thunb. Chem Pharm Bull 54:936–940
Lu HM, Liang YZ, Yi LZ, Wu XJ (2006b) Anti-inflammatory effect of Houttuynia cordata injection. J Ethnopharmacol 104:245–249
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Marles RJ, Farnsworth NR (1995) Antidiabetic plants and their active constituents. Phytomedicine 2:137–189
Mihara T (1960) On the reduction division of Houttuynia cordata Thunb. Bot Mag Tok 73:498–498
Nei M (1972) Genetic distance between populations. Am Nat 106(949):283–292
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genet 89(3):583–590
Peakall R, Smouse PE (2006) GenAlEx 6 - Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinf 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genet 155(2):945–959
Sarkar S (1993) Studies on herbaceous plants of Karbi Anglong District of Assam with reference to their taxonomy and economic utilization. Ph.D. Thesis, Gauhati University, Assam, India
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Mol Ecol 7:465–474
Shibata K, Miyake H (1908) Ueber Parthenogenesis bei Houttuynia cordata. Bot Mag Tok 22:141–144
Shin S, Joo SS, Jeon JH, Park DS, Jang MJ, Kim TO, Kim HK, Hwang BY, Kim KY, Kim YB (2010) Anti-inflammatory effects of a Houttuynia cordata supercritical extract. J Vet Sci 11(3):273–275
Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38:1409–1438
Toda S (2005) Antioxidative effects of polyphenols in leaves of Houttuynia cordata on protein fragmentation by copper-hydrogen peroxide in vitro. J Med Food 8:266–268
Tseng YC (1982) Florae Reipublicae Popularis Sinicae. Science Press, Beijing, p 8
Vekemans X (2002) AFLP-SURV version 1.0. Distributed by the author. Laboratoire de Genetique et Ecologie Vegetale, Universite Libre de Bruxelles, Belgium. Available:http://www.ulb.be/sciences/lagev/aflp-surv.html
Vekemans X, Beauwens T, Lemaire M, Roldan-Ruiz I (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11:139–151
Wei L, Wu X (2012) Genetic variation and population differentiation in a medical herb Houttuynia cordata in China revealed by inter-simple sequence repeats (ISSRs). Int J Mol Sci 13:8159–8170
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Wu W, Zheng Y, Chen L, Yang R, Yan Z, Wei Y (2002) Isozymes variations among the germplasm resources of Houttuynia in Sichuan. J Chin Med Mat 25:695–698
Wu W, Zheng YL, Yang RW, Chen L, Wei YM (2003) Variation of the chromosome number and cytomixis of Houttuynia cordata from China. Acta Phytotaxonomica Sin 41(3):245–257
Wu W, Zheng YL, Chen L, Wei YM, Yang RW, Yan ZH (2005) Evaluation of genetic relationships in the genus Houttuynia Thunb. in China based on RAPD and ISSR markers. Biochem Syst Ecol 33:1141–1157
Yeh FC, Yang RC, Boyle T (1997) POPGENE version 1.31, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Edmonton, Canada. Available: http://www.ualberta.ca/fyeh/index.htm. Accessed 19 Jul 2019
Yin J, Li G, Li J, Yang Q, Ren X (2011) In vitro and in vivo effects of Houttuynia cordata on infectious bronchitis virus. Avian Pathol 40:491–498
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Zhong J, Wang K, Qiu P (2009) Genetic diversity of populations in germplasm resources of Houttuynia cordata Thunb. Acta Agric Boreali-Sin 24:94–100
Zhong J, Wu FC, Qiu P, Dai LJ (2016) Correlation analysis of genetic diversity and population structure of Houttuynia cordata Thunb with regard to environment. Genet Mol Res 15(3)
Zhou NN, Tang J, Chen WD, Feng GK, Xie BF, Liu ZC, Yang D, Zhu XF (2012) Houttuyninum, an active constituent of Chinese herbal medicine, inhibits phosphorylation of HER2/neu receptor tyrosine kinase and the tumor growth of HER2/neu overexpressing cancer cells. Life Sci 90:770–775
Zu YG, Cui JZ (2002) Clonal diversity in Leymus chinensis populations. Acta Phytoecol Sin 26:157–162
Acknowledgements
The authors are thankful to Director, IASST, Paschim Boragaon, Guwahati, Assam, for providing laboratory facilities and Dr. T Bandyopadhyay for proof-reading of the manuscript.
Funding
This work received funding from the Department of Biotechnology, Govt. of India under DBT-RA Scheme.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
• Higher genetic diversity was observed between populations of different regions compared to same region.
• Studied populations are grouped into four clusters according to their geographical regions.
• Low gene flow was observed among studied populations.
Supplementary Information
ESM 1
(DOCX 25 kb)
Rights and permissions
About this article
Cite this article
Gupta, S., Bharalee, R. Genetic Diversity and Population Structure of a Medicinal Herb Houttuynia cordata Thunb. of North-East India. Plant Mol Biol Rep 39, 434–442 (2021). https://doi.org/10.1007/s11105-020-01260-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01260-9