Abstract
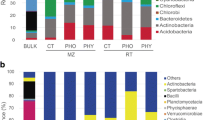
The spatial distribution of alkaline phosphomonoesterase (ALP) activity in the rhizosphere and bulk soil of three plants (Zea mays, Medicago sativa, and Cyperus rotundus) grown in a karst soil (pH 8.27) was visualized using in situ soil zymography. According to the zymogram images, we identified and precisely collected soil samples from hotspots and non-hotspots of ALP activity, and then analyzed the phoD genes that encoded alkaline phosphomonoesterase and assessed the microbial community. The results showed that (1) the phoD abundance in the plant types varied and was highest in the alfalfa and lowest in the maize; (2) Proteobacteria dominated the phoD-harboring microbes in the rhizosphere and Actinobacteria dominated the phoD-harboring microbes in the bulk soil, and (3) the ALP activity was positively correlated with the relative abundance of the diazotrophic Azotobacter, but negatively correlated with the phoD gene abundance, microbial community richness, and diversity. By coupling zymography and microbial molecular approaches, we identified hotspots of enzymatic and microbial activity in rhizosphere soil and evaluated the relative contributions from potential active microorganisms. We found that the function of specific phoD-harboring microorganisms in these hotspots differed depending on the plant in the soil, which had implications for phosphorus (P) management in P-limited karst soils. The results also suggested that free-living N2-fixing bacteria (Azotobacter) might promote ALP activity, thereby emphasizing vital linkages between, and coupling of, the soil nutrient cycles.









Similar content being viewed by others
References
Bargaz A, Lyamlouli K, Chtouki M, Zeroual Y, Dhiba D (2018) Soil microbial resources for improving fertilizers efficiency in an integrated plant nutrient management system. Front Microbiol 9:1606
Beijerinck MW (1913) De Infusies en de ontdekking der backterien, jaarboek van de koninklijke akademie v wetenschappen. Müller, Amsterdam, pp 1–28
Bloom AJ, Chapin FS, Mooney HA (1985) Resource limitation in plants - an economic analogy. Annu Rev Ecol Syst 16:363–392
Burns JA, Zehr JP, Capone DG (2002) Nitrogen-fixing phylotypes of Chesapeake Bay and Neuse River estuary sediments. Microb Ecol 44:336–343
Chen H, Li DJ, Xiao KC, Wang KL (2018) Soil microbial processes and resource limitation in karst and non-karst forests. Funct Ecol 32:1400–1409
Dungait JAJ, Kemmitt SJ, Michallon L, Guo S, Wen Q, Brookes PC, Evershed RP (2013) The variable response of soil microorganisms to trace concentrations of low molecular weight organic substrates of increasing complexity. Soil Biol Biochem 64:57–64
Dynarski KA, Houlton BZ (2018) Nutrient limitation of terrestrial free-living nitrogen fixation. New Phytol 217:1050–1061
Fageria NK, Moreira A (2011) The role of mineral nutrition on root growth of crop plants. Adv Agron 110:251–331
Falkowski PG, Fenchel T, Delong EF (2008) The microbial engines that drive Earth’s biogeochemical cycles. Science 320:1034–1039
FAO, IUSS Working Group WRB (2015) World reference base for soil resources 2014, update 2015: international soil classification system for naming soils and creating legends for soil maps. World Soil Resources Reports No 106, p 192
Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR (2013) FunGene: the functional gene pipeline and repository. Front Microbiol 4:291
Fraser TD, Lynch DH, Bent E, Entz MH, Dunfield KE (2015) Soil bacterial phoD gene abundance and expression in response to applied phosphorus and long-term management. Soil Biol Biochem 88:137–147
Fraser TD, Lynch DH, Gaiero J, Khosla K, Dunfield KE (2017) Quantification of bacterial non-specific acid (phoC) and alkaline (phoD) phosphatase genes in bulk and rhizosphere soil from organically managed soybean fields. Appl Soil Ecol 111:48–56
Giller KE, Chalk P, Dobermann A, Hammond L, Heffer, P, Ladha JK., Nyamudeza P, Maene L, Ssali H, Freney J (2004) Emerging technologies to increase the efficiency of use of fertilzer nitrogen. In: Mosier AR, Syers JK, Freney JR (eds.) Agriculture and the nitrogen cycle: assessing the impacts of fertilizer use on food production and the environment Island Press, Washington DC, pp 35–51
Gillespie AR, Pope PE (1989) Alfalfa N2-fixation enhances the phosphorus uptake of walnut in interplantings. Plant Soil 113:291–293
Gomez PF, Ingram LO (1995) Cloning, sequencing and characterization of the alkaline-phosphatase gene (phoD) from zymomonas-mobilis. Fems Microbiol Lett 125:237–245
Gomola CE, McKay JK, Wallenstein MD, Wagg C, O’Brien MJ (2018) Within-species trade-offs in plant-stimulated soil enzyme activity and growth, flowering, and seed size. Ecol Evol 8:11717–11724
Green SM, Dungait JAJ, Tu CL, Buss HL, Sanderson N, Hawkes SJ, Xing KX, Yue FJ, Hussey VL, Peng J, Johnes P, Barrows T, Hartley IP, Song XW, Jiang ZH, Meersmans J, Zhang XY, Tian J, Wu XF, Liu HY, Song ZL, Evershed R, Gao Y, Quine TA (2019) Soil functions and ecosystem services research in the Chinese karst Critical Zone. Chem Geol 527:119107
Gregory PJ, Bengough AG, George TS, Hallett PD (2013) Rhizosphere engineering by plants: quantifying soil-root interactions. In: Timlin D, Ahuja LR (eds) Enhancing understanding and quantification of soil-root growth interactions. Book Series: advances in agricultural systems modeling-transdisciplinary research synthesis and applications, vol 4. Soil Science Society of America Inc, Madison, pp 1–31
Guber A, Kravchenko A, Razavi BS, Uteau D, Peth S, Blagodatskaya E, Kuzyakov Y (2018) Quantitative soil zymography: mechanisms, processes of substrate and enzyme diffusion in porous media. Soil Biol Biochem 127:156–167
Hinsinger P, Bengough AG, Vetterlein D, Young IM (2009) Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321:117–152
Hu Y, Xia YH, Sun Q, Liu KP, Chen XB, Ge TD, Zhu BL, Zhu ZK, Zhang ZH, Su YR (2018) Effects of long-term fertilization on phoD-harboring bacterial community in Karst soils. Sci Total Environ 628–629:53–63
Iqbal J, Hussain S, Ali A, Javaid A (2012) Biology and management of purple nutsedge (cyperus rotundus l.). J Anim Plant Sci 22:384–389
Iswaran V, Marhwa TS (1982) Nitrogen-fixing bacteria in various tissue-cells of cyperus-rotundus. Zentralblatt Fur Mikrobiologie 137:348–349
Johnston-Monje D, Lundberg DS, Lazarovits G, Reis VM, Raizada MN (2016) Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil 405:337–355
Kpomblekoua K, Tabatabai MA (1994) Effect of organic-acids on release of phosphorus from phosphate rocks. Soil Sci 158:442–453
Kuzyakov Y, Blagodatskaya E (2015) Microbial hotspots and hot moments in soil: concept & review. Soil Biol Biochem 83:184–199
Kuzyakov Y, Razavi BS (2019) Rhizosphere size and shape: temporal dynamics and spatial stationarity. Soil Biol Biochem 135:343–360
LaManna JM, Bothe JV Jr, Zhang FY, Mench MM (2014) Measurement of capillary pressure in fuel cell diffusion media, micro-porous layers, catalyst layers, and interfaces. J Power Sources 271:180–186
Li X, Zeng R, Liao H (2016) Improving crop nutrient efficiency through root architecture modifications Journal of Integrative. Plant Biol 58:193–202
Li DD, Zhang XY, Green SM, Dungait JAJ, Wen XF, Tang YQ, Guo ZM, Yang Y, Sun XM, Quine TA (2018) Nitrogen functional gene activity in soil profiles under progressive vegetative recovery after abandonment of agriculture at the Puding Karst Critical Zone Observatory, SW China. Soil Biol Biochem 125:93–102
Liu S, Razavi BS, Su X, Maharjan M, Zarebanadkouki M, Blagodatskaya E, Kuzyakov Y (2017) Spatio-temporal patterns of enzyme activities after manure application reflect mechanisms of niche differentiation between plants and microorganisms. Soil Biol Biochem 112:100–109
Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA, Doebeli M, Parfrey LW (2018) Function and functional redundancy in microbial systems. Nat Ecol Evol 2:936–943
Luo G, Sun B, Li L, Li MH, Liu MQ, Zhu YY, Guo SW, Ling N, Shen QR (2019) Understanding how long-term organic amendments increase soil phosphatase activities: insight into phoD- and phoC-harboring functional microbial populations. Soil Biol Biochem 139:107632
Ma B, Wang HZ, Dsouza M, Lou J, He Y, Dai ZM, Brookes PC, Xu JM, Gilbert JA (2016) Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J 10:1891–1901
McGuire KL, Treseder KK (2010) Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol Biochem 42:529–535
Nannipieri P, Sequi P, Fusi P (1996) Humus and enzyme activity. In: Piccolo A (ed) Humic substances in terrestrial ecosystems. Elsevier, Amsterdam, pp 293–328
Nannipieri P, Giagnoni L, Landi L, Renella G (2011) Role of phosphatase enzymes in soil. In: Bunemann EK, Oberson A, Frossard E (eds) Phosphorus in action: biological processes in soil phosphorus cycling. Springer, Berlin, pp 215–243
Nannipieri P, Trasar-Cepeda C, Dick RP (2018) Soil enzyme activity: a brief history and biochemistry as a basis for appropriate interpretations and meta-analysis. Biol Fertil Soils 54:11–19
Ng EL, Patti AF, Rose MT, Schefe CR, Wilkinson K, Cavagnaro TR (2014) Functional stoichiometry of soil microbial communities after amendment with stabilised organic matter. Soil Biol Biochem 76:170–178
Oksanen J (2011) Multivariate analysis of ecological communities in R: vegan tutorial. https://john-quensen.com/wp-content/uploads/2018/10/Oksanen-Jari-vegantutor.pdf
Palacios OA, Bashan Y, de-Bashan LE (2014) Proven and potential involvement of vitamins in interactions of plants with plant growth-promoting bacteria-an overview. Biol Fertil Soils 50:415–432
Peerzada AM (2017) Biology, agricultural impact, and management of Cyperus rotundus L.: the world’s most tenacious weed. Acta Physiol Plant 39:270
Postma J, Schilder MT, Bloem J, van Leeuwen-Haagsma WK (2008) Soil suppressiveness and functional diversity of the soil microflora in organic farming systems. Soil Biol Biochem 40:2394–2406
Postma J, Schilder MT, van Hoof RA (2011) Indigenous populations of three closely related Lysobacter spp. in agricultural soils using real-time PCR. Microb Ecol 62:948–958
Ragot SA, Kertesz MA, Buenemann EK (2015) phoD alkaline phosphatase gene diversity in soil. Appl Environ Microbiol 81:7281–7289
Razavi BS, Zarebanadkouki M, Blagodatskaya E, Kuzyakov Y (2016) Rhizosphere shape of lentil and maize: spatial distribution of enzyme activities. Soil Biol Biochem 96:229–237
Razavi BS, Zhang X, Bilyera N, Guber A, Zarebanadkouki M (2019) Soil zymography: simple and reliable? Review of current knowledge and optimization of the method. Rhizosphere 11:100161
Reed SC, Seastedt TR, Mann CM, Suding KN, Townsend AR, Cherwin KL (2007) Phosphorus fertilization stimulates nitrogen fixation and increases inorganic nitrogen concentrations in a restored prairie. Appl Soil Ecol 36:238–242
Reznick D, Bryant MJ, Bashey F (2002) r- and K-selection revisited: the role of population regulation in life-history evolution. Ecology 83:1509–1520
Richardson AE, Simpson RJ (2011) Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol 156:989–996
Sakurai M, Wasaki J, Tomizawa Y, Shinano T, Osaki M (2008) Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Sci Plant Nutr 54:62–71
Salgado TP, Pitelli RA, Alves PLCA, Salvador FL, Nunes AS (2006) Efeitos da adubação fosfatada nas relações de interferência inicial entre plantas de milho (Zea mays) e de tiririca (Cyperus rotundus). Planta Daninha 24:37–44
Salter SJ, Cox MJ, Turek EM, Calus S, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87
Sanaullah M, Razavi BS, Blagodatskaya E, Kuzyakov Y (2016) Spatial distribution and catalytic mechanisms of β-glucosidase activity at the root-soil interface. Biol Fertil Soils 52:505–514
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schoeler A, Jacquiod S, Vestergaard G, Schulz S, Schloter M (2017) Analysis of soil microbial communities based on amplicon sequencing of marker genes. Biol Fertil Soils 53:485–489
Schroeder MS, Janos DP (2005) Plant growth, phosphorus nutrition, and root morphological responses to arbuscular mycorrhizas, phosphorus fertilization, and intraspecific density. Mycorrhiza 15:203–216
Spohn M, Carminati A, Kuzyakov Y (2013) Soil zymography – a novel in situ method for mapping distribution of enzyme activity in soil. Soil Biol Biochem 58:275–280
Vershinina OA, Znamenskaia LV (2002) The Pho regulons of bacteria. Mikrobiologiia 71:581–595
Vestergaard G, Schulz S, Schoeler A, Schloter M (2017) Making big data smart-how to use metagenomics to understand soil quality. Biol Fertil Soils 53:479–484
Wang T, Yang Y, Wenhong MA (2008) Storage, patterns and environmental controls of soil phosphorus in China. Acta Sci Nat Univ Pekin 44:945–952
Wei X, Hu YJ, Razavi BS, Zhou J, Shen JL, Nannipieri P, Wu JS, Ge TD (2019) Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol Biochem 131:62–70
Wright AL, Reddy KR (2001) Phosphorus loading effects on extracellular enzyme activity in everglades wetland soils. Soil Sci Soc Am J 65:588–595
Zhang R, Vivanco JM, Shen Q (2017) The unseen rhizosphere root-soil-microbe interactions for crop production. Curr Opin Microbiol 37:8–14
Funding
This study was supported by the Science Centre project of National Natural Science Foundation of China (Grant Nos. 31988102), and state key and the general programs of the National Natural Science Foundation of China (Grant Nos. 41830860, 41877091, 41571130043), and the National Environmental Research Council of the UK (Grant No. NE/N007603/1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Table S1
The primers used for qPCR and corresponding amplification cycling conditions (DOCX 15 kb)
Fig. S1
Sampling diagram with maize as an example (a) zymogram image showing differences in enzyme activity around the roots. (b) precise sampling hotspots and non-hotspots soils in rhizoboxes. The yellow ellipse represents the bulk soil and the red ellipse represents the rhizosphere (DOCX 2276 kb)
Fig. S2
Distributions of phosphomonoesterase activities in maize, alfalfa, and sedge roots from (a) the root tip upward and (b) the root center outward (DOCX 164 kb)
Fig. S3
DNA concentration (ng g dry soil−1) in the rhizosphere and bulk soils of maize, alfalfa, and sedge rhizoboxes. Upper case letters indicate significant differences (p < 0.05) between different plant species in their rhizosphere and bulk soils, respectively. Lower letters indicate the significant differences (p < 0.05) between soils of the same plant species (DOCX 42 kb)
Rights and permissions
About this article
Cite this article
Liu, S., Zhang, X., Dungait, J.A.J. et al. Rare microbial taxa rather than phoD gene abundance determine hotspots of alkaline phosphomonoesterase activity in the karst rhizosphere soil. Biol Fertil Soils 57, 257–268 (2021). https://doi.org/10.1007/s00374-020-01522-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-020-01522-4