Abstract
Remdesivir was approved by the U.S.A. Food and Drug administration for emergency use to interfere with the replication of SARS CoV-2 virus (the agent that causes COVID-19) in adults and children hospitalized with severe disease. The crystal structure of the metabolite of remdesivir (Monophosphate of GS-441524) and NSP12-NSP8-NSP7 of SARS CoV-2 virus was recently reported. The crystal structures of ADP-Ribose or AMP and NSP3 of SARS CoV-2 virus were also released, recently. This study compared their binding sites and suggests the crystal structure of NSP3 of SARS CoV-2 virus as an alternative binding site of AMP or ADP-ribose to treat COVID-19. We virtually screened 682 FDA-approved compounds, and the top 10 compounds were selected by analysis of docking scores, (G-score, D-score, and Chemscore) and visual analysis using a structure-based docking approach of NSP3 of SARS CoV-2 virus. All immunization approaches are based on the SARS-CoV-2 virus spike protein. A recent study reported that the D614G mutation in the SARS-CoV-2 virus spike protein reduces S1 shedding and increases infectivity of SARS COV-2 virus. Therefore, if there is a severe change in the spike protein of a modified Coronavirus, all developed vaccines can lose their efficacy, necessitating the need for an alternative treatment method. The top 10 compounds (FDA-approved) in this study are selected based on NSP 3 binding site, and therefore are a potential viable treatment because they will show potential activity for all mutations in the SARS-CoV-2 virus spike protein.
Similar content being viewed by others
1 Introduction
Coronaviruses (CoV) were first identified in the 1960s as human pathogens [1]. A new virus, known as the severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 virus) arose in China, rapidly instigating the COVID-19 pandemic. Remdesivir is an antiviral medication developed by Gilead Sciences to treat hepatitis C and was also tested as a potential treatment for Ebola and Coronavirus [2, 3]. Although remdesivir did not originally target SARS CoV-2 virus, remdesivir began to be tested in laboratory by Gilead Sciences in January 2020 and was stated to be active against severe acute respiratory syndrome coronavirus (SARS) and Middle East respiratory syndrome (MERS) in animal models. In 2020, the U.S. Food and Drug Administration issued an emergency use authorization for the investigational antiviral drug remdesivir for the treatment of suspected or laboratory-confirmed SARS CoV-2 virus in adults and children hospitalized with severe disease. The binding mode of remdesivir in SARS CoV-2 virus is not clear, but structures of remdesivir and its parent drug, GS-441524 (Figs. 1 and 2) help understand how they bind in SARS CoV-2 virus and are effective as an antiviral [4]. Recently, Yin reported [5] a structural basis for the inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 virus by remdesivir. The actual binding site of the metabolite of remdesivir (the complex of diphosphate-Mg- monophosphate of GS-441524) is disclosed by Yin, and many crystal structures are released for the AMP (Adenosine monophosphate) or ADP-ribose (Adenosine-5-diphosphoribose) binding site of NSP3 [6, 7]. We observe that the structures of AMP (complex of NSP3) and monophosphate of GS-441524 (complex of NSP12) are very similar. The size of ADP-ribose (complex of NSP3) and the triphosphate of GS-441524 (the active metabolite of remdesivir) [4] are also very similar (ATP-like in Fig. 2); we hypothesize that remdesivir will potentially bind to NSP3 and NSP12-NSP8-NSP7.
Metabolism of remdesivir (prodrug) into GS-41524 (parent drug) [4]
It was reported that the D614G mutation reduces S1 shedding and increases infectivity [8]. All immunization approaches against SARS CoV-2 virus are currently based on this spike protein. Therefore, if there is a severe change in the spike protein of a modified Coronavirus, there is a possibility that the developed vaccines will lose their efficacy. While there is no direct evidence of the effect of the G614 mutant on developing vaccines, this mutation serves as an example of possible future mutations that may cause developing vaccines to lose their efficacy. Unlike immunization approaches, the approach of developing a drug through remdesivir with its NSP binding sites is a promising choice to treat modified SARS CoV-2 viruses.
Clinical trials of remdesivir are still progress, rendering the possible side effects of remdesivir to be unknown. Therefore, in addition to remdesivir, this study searches for alternative treatment against mutations in the spike protein. We perform virtual screening of 682 FDA approved compounds based off the binding modes of remdesivir with NSP3. The top 10 compounds from virtual screening are selected as potential treatments for Coronavirus. We search for additional alternatives in hope that the compounds could potentially test to be more active against Coronavirus in the future.
2 Material and Methods
2.1 General Methods
Visual analysis was performed by observing the hydrogen bonding, molecular surface (MOLCAD) of cavity depth, and lipophilic and electrostatic potentials. The binding cavity was based on each crystal structure (6W6Y, 6WOJ, 6 M71, 7BV2). Superimposition was calculated in “match” function. A commercially available 682 FDA approved data set from Enamine was used for virtual screening through the Surflex-Docking (GEOM) setting. Scoring functions were calculated by CSCORE. All calculations and simulations were done using SYBYL-X2.1 [9].
2.2 Binding Site of NSP3 and NSP12-NSP8-NSP7 of SARS CoV-2 Virus
Two crystal structures were selected for NSP3: AMP (PDB: 6W6Y) and ADP-ribose (PDB: 6WOJ). Using SYBYL-X2.1 [9], structures of the ligand complex were identified, molecular surfaces of ligands were calculated, and H-bonds were analyzed. In addition, the lipophilic and electrostatic interactions were validated for each complex. Two crystal structures (PDB: 6W6Y, 6WOJ) were superimposed and analyzed based on the binding site of AMP and ADP-ribose [6, 7]. An apo structure [10] and a bound structure with the metabolite of remdesivir (monophosphate of GS-441524) [5] were available for NSP12-NSP8-NSP7. Both structures (PDB: 6 M71, 7BV2) were superimposed and compared in the binding site by SYBYL-X2.1.
2.3 Validation of Previously Reported Experimental Data for NSP3 or NSP12 of Coronavirus
This study targets ADP-ribose as a potential binding site as a result of Frick’s report [7] of the molecular basis ADP-ribose binding to the Mac1 domain of SARS CoV-2 virus, NSP3. It was suggested that NSP3 binds to both SARS-CoV virus and SARS-CoV-2 virus, despite the two amino acid sequences varying by 26%. When testing the ADP-ribose binding in the two Coronaviruses, it binds to SARS-CoV-2 virus protein more tightly (Kd = 10 μM) than the SARS-CoV virus protein (Kd = 24 μM). Thus, he encourages the use of ADP-ribose to design antiviral drugs targeting this NSP3 region in SARS CoV-2 virus. This study superimposes the crystal structures of SARS-CoV [11] and SARS CoV-2 viruses using SYBYL-X2.1 to explain the different binding affinities of ADP-ribose to both Coronaviruses. Tchesnokov reported [12] that as shown through stead-state kinetic parameters, the efficiency of ATP incorporation decreases threefold when with the mutant enzyme. To validate these experimental data, the crystal structure of NSP12 (wild type) and V557L were generated and superimposed by SYBYL-X2.1.
2.4 Selected Binding Sites of Virtual Screening in This Study
The crystal structure of NSP3 with ADP-ribose (PDB: 6WOJ) was selected to search potential inhibitors for SARS CoV-2 virus using virtual screening of FDA approved 682 drugs (commercially available from Enamine).
2.5 Selection of Reference Compounds and Top 10 Compounds
Reference compounds are needed for post-docking analysis to select potential compounds for alternative treatments of COVID-19 (via the etiological SARS CoV-2 virus). The reference compounds consisted of structures of ATP, ADP, and AMP-like (size) compounds (all ten compounds from Figs. 1 and 2), along with remdesivir and GS-441524. GS-441524 was added because Eastman and et al. reported how remdesivir is metabolized to GS-441524 [4]. 682 FDA approved compounds were prepared and minimized in MMFF94s (Force Fields) and MMFF94 (Charges) and docked to the NSP3 binding site of the SARS CoV-2 virus crystal structure (PDB: 6WOJ). Scoring functions have been used to estimate and predict which compounds would have the best actual binding affinities in an experimental test. Surflex-score, G-score, D-score, and Chemscore were scoring functions used to predict the best compounds using virtual screening [13,14,15,16,17,18,19]. Although scoring functions are reliable mathematical functions, there is still a possibility for a protein and ligand to clash and protrude [9]. Therefore, visual analysis is crucial in predicting the best potential compounds. Every docked compound was visually analyzed, and compounds with protruding ligands and proteins were eliminated. The scoring functions in this study were compared and analyzed using SYBYL-X2.1 [9] (Table 1).
3 Results
3.1 Binding Site of NSP3, NSP12-NSP8-NSP7
Figures 3 and 4 each display hydrophilic interactions of AMP and ADP-ribose with their hydrogen bonds and NSP3 binding site. Figures 3 and 4 also display that the size of the NSP3 binding site cavity fits well with the AMP and ADP-ribose complex ligands in each crystal structure. The lipophilic and electrostatic molecular surfaces are shown in Figs. 5 and 6 for AMP and ADP-ribose with NSP3 of SARS CoV-2 virus respectively. Both crystal structures (PDB: 6W6Y, 6WOJ) were superimposed and the binding cavities were compared (Fig. 7). Two crystal structures of NSP12-NSP8-NSP7 were superimposed and analyzed (Fig. 8); one is an apo structure [8] (PDB: 6 M71) and another is a bound structure (PDB: 7BV2) with a metabolite of remdesivir (monophosphate of GS-441524) [5].
3.2 Validation of Previously Reported Experimental Data for NSP3 and NSP12 of Coronavirus
The crystal structures of SARS-CoV [11] and SARS CoV-2 viruses were superimposed (PDB id: 6WOJ, 2FAV) in Fig. 9; sequence alignment of two crystal structures of SARS-CoV and SARS CoV-2 viruses was 73.2% in 168 residues. However, in active sites, they were nearly identical, with the exception in only one amino acid of PHE156 for SARS CoV-2, and ASN157 for SARS-CoV virus. Adenosine showed hydrophobic interactions with the phenyl group in PHE156 and had no hydrophobic interactions with ASN157 for SARS-CoV (Fig. 9). It explained why the SARS CoV-2 virus protein binds to ADP-ribose somewhat more tightly (Kd = 10 μM) than the SARS-CoV virus protein (Kd = 24 μM) [7]. The crystal structure of NSP12 (wild type) and its mutation of V557L were generated and superimposed in Fig. 10. The complex of NSP12 and metabolite of remdesivir displayed that it metabolized in monophosphate of GS-441524, bonding with magnesium with diphosphate in Fig. 10. Overall, the size of the structure looks like the main active metabolite of remdesivir, triphosphate of GS-441524, (compound d in Fig. 1) reported by Eastman and Gordon [4, 20]. VAL557 is located in between ARG555 and U10 (Uridine phosphate); the metabolite of remdesivir has H-bonds with ARG555 (NSP12) and U10 (UTP), explaining the counteractive nature of the V557L mutation, in regard to the inhibitory effects of remdesivir; the threefold reduction in the efficiency of ATP incorporation with the mutant enzyme [12]. The H-bond interaction was lost due the H-bond distance between pyrrolo[2,1-f] [1,2,4]triazine, and ARG555 increasing from 2.72 to 3.13 Armstrong (from purple 557 Valine to orange 557 Leucine) in Fig. 10.
3.3 Top 10 Compounds from Virtual Screening of FDA Approved 682 Drugs to NSP3 Binding Site of SARS CoV-2 Virus
Table 2 shows how reference compounds were listed and how the top 10 compounds were selected based on scoring functions [13,14,15,16,17,18,19]. The top 10 compounds, remdesivir, and its parent compound (GS-441524) were each displayed in a docking pose in the cavity depth of molecular surfaces (Fig. 11) and A zoomed out figures of the top 9 compounds were displayed in active sites of amino acids, generating H-binds (Fig. 12) to explain where the ligands are binding relative to the natural substrate (Table 3).
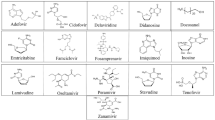
Docking poses in the cavity of NSP3 for top 10 compounds and Redesivir and its parent compound (GS-441524); (a) (top 1) Folic acid, (b) (top 2) Telmisartann, (c) (top 3) Methotrexate, (d) (top 4) Bosentan, (e) (top 5) Lapatinib, (f) (top 6) Gefitinib, (g) (top 7) Ketoconazole, (h) (top 8) Carvedilol, (i) (top 9) Glyburide, (j) (top 10) Avanafil, (k) (refernce) Remdesivir, and (l) (reference) GS-441524
Docking poses in the cavity of NSP3 for top 9 compounds with active sites, generating H-bonds; (a) (top 1) Folic acid, (b) (top 2) Telmisartann, (c) (top 3) Methotrexate, (d) (top 4) Bosentan, (e) (top 5) Lapatinib, (f) (top 6) Gefitinib, (g) (top 7) Ketoconazole, (h) (top 8) Carvedilol, (i) (top 9) Glyburide
4 Discussion
From analyzing the crystal structure of NSP12-NSP7-NSP8, we discovered that the main interaction of remdesivir’s metabolite (monophosphate of GS-441524) was with NSP12. This binding site was superimposed using two available NSP12-NSP7-NSP8 crystal structures. The two structures differ greatly, as one is an apo structure and other is a bound structure, and both are extremely flexible depending on the conditions (Fig. 8). Contrastingly, the superimposed ADP-ribose and AMP (NSP3) binding sites (PDB: 6W6Y, 6WOJ) are visually consistent and stable. Therefore, the NSP3 binding site was selected to virtually screen 692 compounds (10 reference compounds in Table 2 and FDA approved 682 compounds). In this study, the docking scores revealed vitamin B9, Folic acid, as the number one candidate to treat COVID-19 (via SARS CoV-2 virus replication inhibition). The docking pose arranged well with NSP3, and the binding pose looked considerably similar with remdesivir and its metabolite (Fig. 13); Both compounds had Hydrogen bonds with ASP22, PHE132 with NSP3. A strong salt bridge interaction (aspartic acid and amine) in ASP22 was formed for both compounds. It has been reported that maternal folic acid supplementation is commonly used to prevent neural tube defects (NTDs). In addition, it may play a role in preventing pregnancy complications [21,22,23] and increasing anti-HBs titers in the primary response, together with the fact that antibody levels after primary vaccination are strongly associated with the persistence of the protective antibody [24]. Recently Serseg reported that Hispidin and Lepidine E: two natural compounds and folic acid as potential inhibitors of 2019-novel coronavirus main protease (2019-nCoVMpro) using molecular docking and SAR study [25]. It was noted that folic acid is able to inhibit furin, preventing binding by the SARS-CoV-2 virus spike protein, preventing cell entry and virus turnover [26]. In this study by the structure- based approach of NSP3 binding domain, folic acid is a promising candidate to treat and prevent COVID-19 infections. Therefore, folic acid may be a potential treatment for the management of COVID-19.
References
Killerby ME, Biggs HM, Haynes A, Dahl RM, Mustaquim D, Gerber SI, Watson JT (2018) Human coronavirus circulation in the United States 2014–2017. J Clin Virol 101:52–56
Stephens B (2020) The story of remdesivir. The New York Times https://www.nytimes.com/2020/04/17/opinion/remdesivir-coronavirus.html. Accessed 21 Sept 2020
Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, Larson N, Strickley R, Wells J, Stuthman KS, Van Tongeren SA, Garza NL, Donnelly G, Shurtleff AC, Retterer CJ, Gharaibeh D, Zamani R, Kenny T, Eaton BP, Grimes E, Welch LS, Gomba L, Wilhelmsen CL, Nichols DK, Nuss JE, Nagle ER, Kugelman JR, Palacios G, Doerffler E, Neville S, Carra E, Clarke MO, Zhang L, Lew W, Ross B, Wang Q, Chun K, Wolfe L, Babusis D, Park Y, Stray KM, Trancheva I, Feng JY, Barauskas O, Xu Y, Wong P, Braun MR, Flint M, McMullan LK, Chen SS, Fearns R, Swaminathan S, Mayers DL, Spiropoulou CF, Lee WA, Nichol ST, Cihlar T, Bavari S (2016) Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 7594:381–385
Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, Hall MD (2020) Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of SARS COV-2. ACS Cent Sci 6:672–683
Yin W, Mao C, Luan X, Hou F, Zhao W, Gao M, Chang S, Xie Y, Tian G, Jiang H, Tao S, Shen J, Jiang Y, Jiang H, Xu Y, Zhang S, Zhang Y, Xu HE (2020) Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368:1499–1504
Michalska K, Kim Y, Jedrzejczak R, Maltseva NI, Stols L, Endres M, Joachimiak A (2020) Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ 7:814–824
Frick DN, Virdi RS, Vuksanovic N, Dahal N, Silvaggi NR (2020) Molecular basis for ADP-ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry 59:2608–2615
Fernández A (2020) Structural impact of mutation D614G in SARS-CoV-2 spike protein: enhanced infectivity and therapeutic opportunity. ACS Med Chem Lett 11:1667–1670
Forino M, Jung D, Easton JB, Houghton PJ, Pellecchia M (2005) Virtual docking approaches to protein kinase B inhibition. J Med Chem 48:2278–2281
Gao Y, Yan L, Huang Y, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z (2020) Structure of the RNA-dependent RNA polymerase from SARS COV-2 virus. Science 368:779–782
Egloff MP, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, Ahola T, Canard B (2006) Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80:8493–8502
Tchesnokov EP, Gordon CJ, Woolner E, Kocincova D, Perry JK, Feng JY, Porter DP, Gotte M (2020) Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J Biol Chem. jbc.AC120.015720. https://doi.org/10.1074/jbc.AC120.015720
Guedes IA, Pereira FSS, Dardenne LE (2018) Empirical scoring functions for structure-based virtual screening: applications, critical aspects, and challenges. Front Pharmacol 9:1089–1106
Li Y, Han L, Liu Z, Wang R (2014) Comparative assessment of scoring functions on an updated benchmark: 2. Evaluation methods and general results. J Chem Inf Model 54:1717–1736
Jain AN (2003) Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 46:499–511
Mooij WT, Verdonk ML (2005) General and targeted statistical potentials for protein-ligand interactions. Proteins 61:272–287
Muegge I (2006) PMF scoring revisited. J Med Chem 49:5895–5902
Ewing TJ, Makino S, Skillman AG, Kuntz ID (2001) DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des 15:411–428
Baxter CA, Murray CW, Clark DE, Westhead DR, Eldridge MD (1998) Flexible docking using Tabu search and an empirical estimate of binding affinity. Proteins 33:367–382
Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, Götte M (2020) Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 295:6785–6797
Zhao X, Pang X, Wang F, Cui F, Wang L, Zhang W (2018) Maternal folic acid supplementation and antibody persistence 5 years after hepatitis B vaccination among infants. Hum Vaccin Immunother 14:2478–2484
Catov JM, Bodnar LM, Olsen J, Olsen S, Nohr EA (2011) Periconceptional multivitamin use and risk of preterm or small-for-gestational-age births in the Danish National Birth Cohort. Am J Clin Nutr 94:906–912
Saccone G, Sarno L, Roman A, Donadono V, Maruotti GM, Martinelli P (2016) 5-Methyl-tetrahydrofolate in prevention of recurrent preeclampsia. J Matern Fetal Neonatal Med 29:916–920
Schonberger K, Riedel C, Ruckinger S, Mansmann U, Jilg W, Kries RV (2013) Determinants of long-term protection after hepatitis B vaccination in infancy: a meta-analysis. Pediatr Infect Dis J 32:307–313
Serseg T, Benarous K, Yousfi M (2020) Hispidin and Lepidine E: two natural compounds and folic acid as potential inhibitors of 2019-novel coronavirus main protease (2019-nCoVMpro), molecular docking and SAR study. Curr Comput Aided Drug Des. https://doi.org/10.2174/1573409916666200422075440
Shakoor H, Feehan J, Mikkelsen K, Al Dhaheri AS, Ali HI, Platat C, Ismail LC, Stojanovska L, Apostolopoulos V (2020) Be well: a potential role for vitamin B in COVID-19. Maturitas S0378-5122(20):30348-0. https://doi.org/10.1016/j.maturitas.2020.08.007
Author information
Authors and Affiliations
Contributions
LSJ, TMG, and MN conceptualized and designed the experiments and analyzed the data. MN reviewed the data and manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jung, L.S., Gund, T.M. & Narayan, M. Comparison of Binding Site of Remdesivir and Its Metabolites with NSP12-NSP7-NSP8, and NSP3 of SARS CoV-2 Virus and Alternative Potential Drugs for COVID-19 Treatment. Protein J 39, 619–630 (2020). https://doi.org/10.1007/s10930-020-09942-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09942-9