Abstract
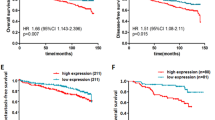
Our recent research has revealed that passenger strands of certain microRNAs (miRNAs) function as tumor-suppressive miRNAs in cancer cells, e.g., miR-101-5p, miR-143-5p, miR-144-5p, miR-145-3p, and miR-150-3p. Thus, they are important in cancer pathogenesis. Analysis of the miRNA expression signature of breast cancer (BrCa) showed that the expression levels of two miRNAs derived from pre-miR-99a (miR-99a-5p and miR-99a-3p) were suppressed in cancerous tissues. The aim of this study was to identify oncogenic genes controlled by pre-miR-99a that are closely involved in the molecular pathogenesis of BrCa. A total of 113 genes were identified as targets of pre-miR-99a regulation (19 genes modulated by miR-99a-5p, and 95 genes regulated by miR-99a-3p) in BrCa cells. Notably, FAM64A was targeted by both of the miRNAs. Among these targets, high expression of 16 genes (C5orf22, YOD1, SLBP, F11R, C12orf49, SRPK1, ZNF250, ZNF695, CDK1, DNMT3B, TRIM25, MCM4, CDKN3, PRPS, FAM64A, and DESI2) significantly predicted reduced survival of BrCa patients based upon The Cancer Genome Atlas (TCGA) database. In this study, we focused on FAM64A and investigated the relationship between FAM64A expression and molecular pathogenesis of BrCa subtypes. The upregulation of FAM64A was confirmed in BrCa clinical specimens. Importantly, the expression of FAM64A significantly differed between patients with Luminal-A and Luminal-B subtypes. Our data strongly suggest that the aberrant expression of FAM64A is involved in the malignant transformation of BrCa. Our miRNA-based approaches (identification of tumor-suppressive miRNAs and their controlled targets) will provide novel information regarding the molecular pathogenesis of BrCa.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Economopoulou P, Dimitriadis G, Psyrri A. Beyond BRCA: new hereditary breast cancer susceptibility genes. Cancer Treat Rev. 2015;41:1–8.
Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–20.
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–16.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. and Panel Members. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the primary therapy of early breast cancer. Ann Oncol. 2011;22:1736–47.
Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Prim. 2019;5:66.
Rakha EA, Fresia GP. New advances in molecular breast cancer pathology. Semin Cancer Biol. 2020;S1044-579X:30080–8.
Jennifer JG, Sandra MS. Luminal a breast cancer and molecular assays: a review. Oncologist. 2018;23:556–65.
Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs—an update. Nat Rev Clin Oncol. 2018;15:541–63.
Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24.
Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37.
Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22.
Toda H, Kurozumi S, Kijima Y, Idichi T, Shinden Y, Yamada Y, et al. Molecular pathogenesis of triple-negative breast cancer based on microRNA expression signatures: antitumor miR-204-5p targets AP1S3. J Hum Genet. 2018;63:1197–210.
Toda H, Seki N, Kurozumi S, Shinden Y, Yamada Y, Nohata N, et al. RNA-sequence-based microRNA expression signature in breast cancer: tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol Oncol. 2020;14:426–46.
Osako Y, Seki N, Koshizuka K, Okato A, Idichi T, Arai T, et al. Regulation of SPOCK1 by dual strands of pre-miR-150 inhibit cancer cell migration and invasion in esophageal squamous cell carcinoma. J Hum Genet. 2017;62:935–44.
Misono S, Seki N, Mizuno K, Yamada Y, Uchida A, Arai T, et al. Dual strands of the miR-145 duplex (miR-145-5p and miR-145-3p) regulate oncogenes in lung adenocarcinoma pathogenesis. J Hum Genet. 2018;63:1015–28.
Uchida A, Seki N, Mizuno K, Misono S, Yamada Y, Kikkawa N, et al. Involvement of dual-strand of the miR-144 duplex and their targets in the pathogenesis of lung squamous cell carcinoma. Cancer Sci. 2019;110:420–32.
Wada M, Goto Y, Tanaka T, Okada R, Moriya S, Idichi T, et al. RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation. J Hum Genet. 2020;65:1019–34.
Mitra R, Adams CM, Jiang W, Greenawalt E, Eischen CM. Pan-cancer analysis reveals cooperativity of bothstrands of microRNA that regulate tumorigenesis and patient survival. Nat Commun. 2020;11:968.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Pereira B, Chin S, Rueda OM, Moen VH, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Jiang L, Ren L, Zhang X, Chen H, Chen X, Lin C, et al. Overexpression of PIMREG promotes breast cancer aggressiveness via constitutive activation of NF-κB signaling. EBioMedicine. 2019;43:188–200.
Yonemori K, Seki N, Idichi T, Kurahara H, Osako Y, Koshizuka K, et al. The microRNA expression signature of pancreatic ductal adenocarcinoma by RNA sequencing: anti-tumour functions of the microRNA-216 cluster. Oncotarget. 2017;8:70097–115.
Koshizuka K, Nohata N, Hanazawa T, Kikkawa N, Arai T, Okato A, et al. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget. 2017;8:30288–304.
Hu Y, Zhu Q, Tang L. MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS ONE. 2014;9:e92099.
Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47:587–95.
Wang X, Li Y, Qi W, Zhang N, Sun M, Huo Q, et al. MicroRNA-99a inhibits tumor aggressive phenotypes through regulating HOXA1 in breast cancer cells. Oncotarget. 2015;6:32737–47.
Xia M, Li H, Wang JJ, Zeng HJ, Wang SH. MiR-99a suppress proliferation, migration and invasion through regulating insulin-like growth factor 1 receptor in breast cancer. Eur Rev Med Pharmacol Sci. 2016;20:1755–63.
Qin H, Liu W. MicroRNA-99a-5p suppresses breast cancer progression and cell-cycle pathway through downregulating CDC25A. J Cell Physiol. 2019;234:3526–37.
Long X, Shi Y, Ye P, Guo J, Zhou Q, Tang Y, et al. MicroRNA-99a suppresses breast cancer progression by targeting FGFR3. Front Oncol. 2020;9:1473.
Osako Y, Yoshino H, Sakaguchi T, Sugita S, Yonemori M, Nakagawa M, et al. Potential tumor-suppressive role of microRNA-99a-3p in sunitinib-resistant renal cell carcinoma cells through the regulation of RRM2. Int J Oncol. 2019;54:1759–70.
Arai T, Okato A, Yamada Y, Sugawara S, Kurozumi A, Kojima S, et al. Regulation of NCAPG by miR-99a-3p (passenger strand) inhibits cancer cell aggressiveness and is involved in CRPC. Cancer Med. 2018;7:1988–2002.
Okada R, Koshizuka K, Yamada Y, Moriya S, Kikkawa N, Kinoshita T, et al. Regulation of oncogenic targets by miR-99a-3p (passenger strand of miR-99a-duplex) in head and neck squamous cell carcinoma. Cells. 2019;8:1535.
Leech AO, Vellanki SH, Rutherford EJ, Keogh A, Jahns H, Hudson L, et al. Cleavage of the extracellular domain of junctional adhesion molecule-A is associated with resistance to anti-HER2 therapies in breast cancer settings. Breast Cancer Res. 2018;20:140.
So JY, Skrypek N, Yang HH, Merchant AS, Nelson GW, Chen WD, et al. Induction of DNMT3B by PGE2 and IL6 at distant metastatic sites promotes epigenetic modification and breast cancer colonization. Cancer Res. 2020;80:2612–27.
Walsh LA, Alvarez MJ, Sabio EY, Reyngold M, Makarov V, Mukherjee S, et al. An integrated systems biology approach identifies TRIM25 as a key determinant of breast cancer metastasis. Cell Rep. 2017;20:1623–40.
Archangelo LF, Gläsner J, Krause A, Bohlander SK. The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10. Oncogene. 2006;25:4099–109.
Archangelo LF, Greif PA, Hölzel M, Harasim T, Kremmer E, Przemeck GK, et al. The CALM and CALM/AF10 interactor CATS is a marker for proliferation. Mol Oncol. 2008;2:356–67.
Caudell D, Aplan PD. The role of CALM-AF10 gene fusion in acute leukemia. Leukemia. 2008;22:678–85.
Hu S, Yuan H, Li Z, Zhang J, Wu J, Chen Y, et al. Transcriptional response profiles of paired tumor-normal samples offer novel perspectives in pan-cancer analysis. Oncotarget. 2017;8:41334–47.
Yao Z, Zheng X, Lu S, He Z, Miao Y, Huang H, et al. Knockdown of FAM64A suppresses proliferation and migration of breast cancer cells. Breast Cancer. 2019;26:835–45.
Zhang J, Qian L, Wu J, Lu D, Yuan H, Li W, et al. Up-regulation of FAM64A promotes epithelial-to-mesenchymal transition and enhances stemness features in breast cancer cells. Biochem Biophys Res Commun. 2019;513:472–8.
Ignatiadis M, Sotiriou C. Luminal breast cancer: from biology to treatment. Nat Rev Clin Oncol. 2013;10:494–506.
Gao JJ, Swain SM. Luminal A breast cancer and molecular assays: a review. Oncologist. 2018;23:556–65.
Kyrochristos ID, Ziogas DE, Roukos DH. Dynamic genome and transcriptional network-based biomarkers and drugs: precision in breast cancer therapy. Med Res Rev. 2019;39:1205–27.
Araki K, Miyoshi Y. Mechanism of resistance to endocrine therapy in breast cancer: the important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer. 2018;25:392–401.
O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13:417–30.
Sobhani N, D’Angelo A, Pittacolo M, Roviello G, Miccoli A, Corona SP, et al. Updates on the CDK4/6 inhibitory strategy and combinations in breast. Cancer Cells. 2019;8:321.
Pandey K, An HJ, Kim SK, Lee SA, Kim S, Lim SM, et al. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: a review. Int J Cancer. 2019;145:1179–88.
Acknowledgements
The present study was supported by KAKENHI grants 18K09338, 19K09049, and 19K18060.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
NN is an employee of MSD K.K., a subsidiary of Merck & Co., Inc. and reports personal fees from MSD K.K. outside this study. The authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shinden, Y., Hirashima, T., Nohata, N. et al. Molecular pathogenesis of breast cancer: impact of miR-99a-5p and miR-99a-3p regulation on oncogenic genes. J Hum Genet 66, 519–534 (2021). https://doi.org/10.1038/s10038-020-00865-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-00865-y